Abstract
Background
The vertebrate opsins are proteins which utilise a retinaldehyde chromophore in their photosensory or photoisomerase roles in the visual/irradiance detection cycle. The majority of the opsins, such as rod and cone opsins, have a very highly conserved gene structure suggesting a common lineage. Exceptions to this are RGR-opsin and melanopsin, whose genes have very different intron insertion positions. The gene structure of another opsin, peropsin (retinal pigment epithelium-derived rhodopsin homologue, RRH) is unknown.
Results
By in silico analysis of the GenBank database we have determined that the human RRH comprises 7 exons spanning approximately 16.5 kb and is localised to chromosome 4q25 in the following gene sequence: cen-EGF-RRH-IF-qter – a position that excludes this gene as a candidate for the RP29 autosomal recessive retinitis pigmentosa locus. A comparison of opsin gene structures reveals that RRH and RGR share two common intron (introns 1 and 4) insertion positions which may reflect a shared ancestral gene.
Conclusion
The opsins comprise a diverse group of genes which appear to have arisen from three different lineages. These lineages comprise the "classical opsin superfamily" which includes the rod and cone opsins, pinopsin, VA-opsin, parapinopsin and encephalopsin; the RRH and RGR group; and the melanopsin line. A common lineage for RRH and RGR, together with their sites of expression in the RPE, indicates that peropsin may act as a retinal isomerase.
Similar content being viewed by others
Background
The photosensory opsins are a family of membrane bound, heptahelical G-protein coupled receptors (GPCRs) characterised by their ability to covalently bind a vitamin A-based retinaldehyde chromophore via a Schiff base to a lysine residue located in the 7th transmembrane α-helix [1, 2]. In the natural environment, the vertebrate vitamin A based chromophore takes the form of either 11-cis-retinal (A1) or 11-cis-3,4-didehydroretinal (A2) [3]. In the vertebrate retina, the primary events in image detection occur in the outer segments of rod and cone cells, with the opsins located in these lamellar stacks being named after these cell types – rod-opsin in rods and cone-opsins in cones. Absorption of a photon of light by the chromophore located in the retinal binding pocket of an opsin causes its photoisomerisation from the 11-cis to an all-trans conformation. This change of the chromophore induces a conformational change of its surrounding opsin molecule allowing transducin (G-protein) binding and activation of the phototransduction cascade [1, 2]. The vertebrate retina generally contains a single rod opsin class and as many as four different cone opsins [4], though the majority of mammals are dichromats being in possession of only two types of cone opsin. In the non-mammals, non-rod, non-cone photosensory opsins have been described. For example, pinopsin (P-opsin) was isolated from the avian pineal [5] and vertebrate ancient opsin (VA-opsin) was isolated from the teleost inner retina and pineal/deep brain structures [6]. Other opsins have also been described from a variety of vertebrate classes which either act as a photoisomerase, e.g. RGR-opsin [7]; or have an unknown function, e.g. peropsin [8], parapinopsin [9] and encephalopsin [10]; or are expressed in photoreceptive cells but have as yet an unknown function, e.g. melanopsin [11]. The one key feature present in all the opsin classes is the presence of a lysine residue in the 7th transmembrane α-helix which is thought to enable retinal attachment in all cases [12].
The first complete vertebrate opsin sequence (bovine rod opsin) was described in 1983 [13], and this opsin has since become the numeration template for critical residues that are conserved throughout all of the opsins. The genomic structure of bovine rod opsin consists of five exons separated by four introns [13], a structure that has been conserved in all vertebrate rod-opsins including those of the ancient Agnatha [14], with the single exception of the rod opsin in teleost fish which is intronless [15]. Employing the nomenclature proposed by Hunt et al. [16] which correlates the spectral sensitivity (λmax) with amino acid sequence identity, the first cone-opsin sequences isolated were those of the human blue (ultraviolet/violet sensitive, UVS/VS) (OPN1SW), green and red cone opsins (both longwave sensitive, LWS) (OPN1MW, OPN1LW) [17]. The UVS/VS opsin gene shares the same genomic structure as the rod-opsin gene in that is possesses four conserved intron insertion positions. The LWS opsin also shares the four conserved introns with the rod- and UVS/VS cone-opsin, but has an additional fifth intron in a 5' prime position to the four conserved introns [17], which we have termed intron 0 (zero) [12]. Sequences from the remaining two visual opsin classes of the vertebrates, shortwave sensitive (SWS) and middlewave sensitive (MWS), have been isolated and share the same genomic structure as the rod and UVS/VS cone opsins [18]. Interestingly, the SWS and MWS opsin families found in birds, fish, reptiles and amphibia are absent in the mammalian lineage – a feature thought to be related to the "nocturnal bottleneck" occupied by mammals during their evolution resulting in the loss of these two cone classes [19, 20].
Many of the more recently described non-rod, non-cone opsins also share a relatively high degree of intron position conservation with the visual opsins, only differing in the position of intron 2 as in pinopsin where it is shifted 14 nucleotides in a 3' direction [21] and VA-opsin where it is shifted 42 nucleotides in a 3' direction [22], or its absence as in parapinopsin [9] and encephalopsin (OPN3) [23]. However, RGR-opsin (retinal G-protein coupled receptor, RGR) [24] and melanopsin (OPN4) [25] do not share intron insertion positions with any other opsins for which a gene structure is known, whilst the gene structure of peropsin is presently unknown.
Little is known about peropsin (retinal pigment epithelium-derived rhodopsin homologue, RRH) except that it shares ~26% amino acid identity with the photosensory opsins, is uniquely expressed in the retinal pigment epithelium (RPE), and by synteny with mouse will map to human chromosome 4q [8]. To address this issue, we have determined by in silico database searches that the human peropsin gene (RRH) has seven exons and maps to chromosome 4q25. A comparison with the gene structures of other opsins indicates that RRH and RGR share two intron insertion sites. These findings and their phylogenetic relationships are discussed within the context of the possible function of peropsin. In view of the variety of nomenclature systems that are applied to the various opsin classes [2, 12], we have attempted to be as consistent as possible with nomenclature and have deferred to preferred locus symbols accepted by the Gene Nomenclature Committee of The Human Genome Organisation, HUGO – http://www.hugo-international.org/hugo/ and Online Mendelian Inheritance in Man, OMIM – http://www.ncbi.nlm.nih.gov/omim/. Thus, RRH and peropsin should be considered synonymous.
Results and Discussion
Structure of the RRH gene
We have used the 1374 base pairs (bp) of the RRH (peropsin) cDNA sequence (GenBank: AF012270) that has been previously described [8] to conduct an online BLAST search of the GenBank nr database. The RRH cDNA matched 7 non-contiguous regions in the Plus/Plus strand orientation of the 118642 bp human BAC clone RP11-602N24 (GenBank: AC126283) at a level of 99–100% identity with highly significant Expect-values (E-value) – the closer the value to "0" the more "significant" the match [26]. Subsequently, a BLAST 2 Sequences comparison of RP11-602N24 with the RRH cDNA indicated that the RRH cDNA is contained within 7 putative exons. Using these putative exons as a model, a sequence alignment was produced such that each intron concurred with the GT/AG intron donor/acceptor site rule [27] (Figure 1). Exon 1 contains the bases 17–156 of the RRH cDNA, which are equivalent to 34 bp of 5' untranslated region (UTR) and the initial 106 bp of coding sequence. We were unable to align the first 16 bp of the RRH cDNA to RP11-602N24 and have concluded that these 16 bp are a cDNA cloning artifact since the breakdown of sequence similarity does not occur at an apparent intron acceptor site in the genomic sequence. Exons 2–6 contain the next 793 bp of coding sequence, whilst exon 7 contains the final 115 bp of coding sequence and the 3'UTR. No nucleotide substitutions were observed between the published coding sequence and that derived from RP11-602N24, but three separate nucleotide substitutions were observed in the 3'UTR. The entire exon/intron structure encompasses approximately 16.5 kilo base pairs (kb) and has been ascribed GenBank accession number BK000958.
Chromosomal localisation of RRH
In order to map RRH, we subjected the sequence of RP11-602N24 to a BLAST search and found that it contains the I factor (complement) gene, IF, which has been shown to map to 4q25 [28]. Localisation of RP11-602N24 and hence RRH to 4q25 is consistent with the syntenic prediction from the mapping data for mouse Rrh [8]. To further refine the localisation of RRH, a BLAST search using a terminal 2 kb of sequence (bases 116643–118642) of RP11-602N24 identified BAC clone B200N5 which maps to 4q25 and contains approximately 143 kb of sequence (GenBank: AC005509). A BLAST search using B200N5 indicated that this clone contains the epidermal growth factor gene, EGF, which is also known to map to 4q25 in the order cen-EGF-IF-qter [28]. We assembled a mini-contig consisting of approximately 260 kb of sequence from clones RP11-602N24 and B200N5 and have been able to determine the following gene order: cen-EGF~85 kb~RRH~26 kb~IF-qter, and their directions of transcription (Figure 2). Localisation of RRH to an interval of approximately 110 kb between EGF and IF on chromosome 4q25 excludes this gene as a causative candidate for the autosomal recessive retinitis pigmentosa locus (RP29) located on 4q32-q34 [29].
Mini-contig showing the arrangement of genes around the RRH locus. Clone names (with associated GenBank accession numbers) are given as are their sizes and regions of overlap in bp. The direction of transcription and location of translation initiation codons (atg) of the various genes are also indicated in bp.
Comparison of opsin gene structures
To determine whether any of the intron insertion sites in RRH are conserved with those of other opsins, nucleotide sequences of representatives of the various opsin classes were aligned and intron positions marked (Figure 3). RRH shares two common intron insertion sites (1 and 4) with RGR, whilst the other four intron insertion sites appear novel amongst the opsin families represented. RRH and RGR share two further introns that are present in relatively close proximity. Intron 3 of RRH is located 9 bp in a 3' direction of intron 3 in RGR (both introns being in phase +1), whilst intron 5 in RRH is located 3 bp (1 codon) in a 3' direction of the corresponding intron in RGR (both introns being in phase 0). Between RRH and members of the other opsin families only two intron positions are in relatively close proximity – intron 6 in RRH is located 1 bp 5' of intron 4 in the rod and cone opsins, and intron 2 of RRH (phase 0) is 8 bp 3' of intron 3 (phase +1) of OPN4.
Human RRH coding sequence and conceptual translation (peropsin) with transmembrane domains (boldface) as predicted by the rod opsin model of Palczewski et al. [48]. The six intron insertion sites in the RRH gene defined in Figure 1 are indicated by black filled circles. Equivalent intron insertion sites for the visual (rod and cone) opsins are indicated by red filled circles – intron positions 1–4 are common to all rod (RHO) and cone opsins, whilst intron 0 is only found in the LWS opsins (OPN1MW, OPN1LW) [13, 17]. The shifted second intron of the pinopsin and VA-opsin families [21, 22] are indicated as 2a and 2b respectively in red circles. Parapineal opsin [9] and encephalopsin (OPN3) [10] possess only introns 1, 3 and 4 of the rod and cone opsins. Equivalent intron insertion sites for RGR-opsin (RGR) [24] are indicated by open circles, whilst those for melanopsin (OPN4) [25] are indicated by yellow filled circles. Only intron insertion sites 1–7 for melanopsin are indicated due to the extreme length of the melanopsin C-terminus which makes positioning of introns 8 and 9 inaccurate since there is no overlap with any other opsin. Note that intron insertion sites 1 and 4 in RRH are equivalent to intron insertion sites 1 and 4 in RGR.
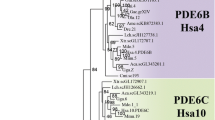
From the perspective of intron positions, it would appear that the opsins have arisen from three ancestral genes (Figure 4). Considering the clade (81% bootstrap confidence) which includes the visual opsins (rod and cones), brain (pinopsin, VA-opsin, parapinopsin) and encephalopsin, it can be seen that three intron positions (introns 1, 3 and 4) are perfectly conserved throughout (Figures 3 and 4). This would suggest that the ancestral opsin gene giving rise to this clade had introns present at these three positions. Subsequent gene duplication events have given rise to opsin classes that either do not possess intron 2, or possess intron 2 in at least one of three positions (position 2, 2a or 2b), whilst the LWS cone opsin group has accumulated a further intron (0) in the visual opsin lineage (Figure 4). Given the organisation of the lamprey (Agnatha) rod opsin gene [14] and pinopsin gene [30] (which has an intron arrangement of the VA-opsin family rather than that of the pinopsin family [22]), these highly conserved gene structures have been in existence for at least 550 million years. Further support for an ancestral chordate opsin gene with these three conserved introns is provided by an opsin from the sea squirt Ciona intestinalis. This urochordate possesses Ci-opsin1 whose gene has seven introns, three of which are in positions conserved with those in vertebrate visual opsins [31], corresponding to positions 1, 3 and 4 [12]. Given their common lineage, we have chosen to term this collection of opsins the "classical opsin superfamily".
Maximum parsimony tree with bootstrap confidence levels based on nucleotide coding sequence of the various vertebrate opsin classes rooted with human melanopsin (see Materials and Methods). Cone opsin class nomenclature after Hunt et al. [16]. To the right of the tree is a schematic representation of the intron insertion sites in the various opsin genes based on Figure 3. Conserved intron positions between opsin classes are indicated by a dashed vertical line. The relative position of the second rod and cone opsin intron is indicated by an arrow head in the pinopsin and VA-opsin classes to show their shifted second intron positions (2a and 2b).
RRH and RGR form a separate clade (87% bootstrap confidence) and share two conserved intron positions with each other, 1 and 4, but none with classical opsin superfamily or melanopsin (Figures 3 and 4). This may be indicative that RRH and RGR derive from the same ancestral gene which possessed introns at positions equivalent to positions 1 and 4 in RRH and RGR, even though they only share ~25% amino acid identity across the transmembrane domains compared with ≥ 40% identity exhibited between the majority of the classical opsin superfamily [12]. An exception being encephalopsin which shares ~30% identity with other members of the classical opsin superfamily [12], but clearly has a highly conserved gene structure. The phenomenon of intron sliding or slippage [32], which has been proposed as a mechanism contributing to the slight variations in position of some introns observed between some insect opsins [33], may explain the 1 bp shift between intron 6 of RRH and intron 4 of the classical opsin superfamily, but their proximity may also be due to chance. Whilst single nucleotide intron slippage is an evolutionary phenomenon thought to occur in <5% of introns [34], these two opsin groups do not appear to have a recent common ancestor which probably indicates a random insertion of introns. Similarly, the close proximity of introns 2 and 3 of RRH with introns 3 of OPN4 and RGR respectively are likely also to be chance occurrences given that intron insertion and deletion events are much more common than intron sliding events [32, 35, 36].
OPN4 (melanopsin) shares no conserved intron positions with any of the other opsin classes, and it has been proposed that this opsin class represents a separate line of opsin evolution [12, 37]. Collectively the data suggest that three separate lines of evolution have occurred to form the present day vertebrate opsins represented by the structurally highly conserved classical opsin superfamily, the more relaxed RRH and RGR grouping, and OPN4 which stands as the single member of the melanopsin class to date.
An ancestral link between RRH and RGR may provide an insight at to the potential function of peropsin. RGR-opsin is known to act as a photoisomerase in the reconversion of retinal chromophore from the all-trans to the 11-cis conformation [38, 39], and perhaps peropsin also functions as an isomerase in some manner given its localisation in the RPE. This suggestion is supported by recent findings that indicate that an amphioxus (Branchiostoma belcheri) homologue of peropsin acts as an all-trans to 11-cis photoisomerase [40]. Alternatively, peropsin may act as a thermal isomerase since Rgr-/- mice are able to convert all-trans-retinal to the 11-cis conformation under dark adaptation [7]. It has been suggested that peropsin may also bind a different retinal isomer, or indeed a non-retinoid ligand [8]. We have recently shown that the non-rod, non-cone opsins such as RGR-opsin, peropsin, melanopsin and encephalopsin are all expressed early in the embryonic development of the mammalian eye. These opsins are expressed by E11.5 in mice and by 8.6 weeks in humans [41], a finding which in all cases predates the expression of the visual rod and cone opsins [42]. This early expression pattern may be supportive of a role in retinoid metabolism for these opsins in the developing retina [43], or that these opsins have a role in the embryonic eye that are quite different to those in the adult eye.
Conclusions
Using GenBank database searches we have been able to determine that the human peropsin gene, RRH, comprises seven exons spanning about 16.5 kb. By assembling sequences from large genomic clones we have determined that RRH localises within an interval of approximately 110 kb between EGF and IF on chromosome 4q25. Of the six introns present in RRH, two are located in positions conserved with introns 1 and 4 of RGR, which given the phylogenetic relationship of RRH and RGR may suggest that these two opsins have arisen from a common ancestor, and by inference possess a common function e.g. acting as a retinal chromophore (photo)isomerase. Our data also strengthens the argument that the present day opsins are represented by genes that have arisen from three different ancestral genes that have given rise to (1), a classical opsin superfamily cosisting of visual opsins and those opsins who share a highly conserved gene structure with the visual opsins; (2), a peropsin and RGR-opsin family; (3), a melanopsin family.
Methods
Database Searches
The GenBank database was screened using the online BLAST [26] server – http://www.ncbi.nlm.nih.gov/BLAST/. Searches were carried out using the standard nucleotide-nucleotide BLAST (blastn) option against the "nr database" using default values with the low complexity filter off. Subsequent sequence manipulations utilised the online BLAST 2 Sequences [44] server – http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html and MacVector 7.0 (Accelrys Ltd., Cambridge, UK).
Phylogenetic Analysis
Nucleotide coding sequences of: (i) human rod opsin, RHO (U49742); (ii) human red cone (LWS) opsin, OPN1LW (AH005298); (iii) human blue cone (UVS/VS) opsin, OPN1SW (U53874); (iv) chicken green cone (MWS) opsin (M92038); (v) chicken blue cone (SWS) opsin (M92037); (vi) chicken pinopsin (U15762); (vii) carp VA-opsin (AF233520); (viii) catfish parapinopsin (AF028014); (ix) human encephalopsin, OPN3 (AF140242); (x) human peropsin, RRH (AF012270); (xi) human RGR-opsin, RGR (U14910); (xii) human melanopsin, OPN4 (AF147788), were entered in to DAMBE version 4.1.15 [45] – http://aix1.uottawa.ca/~xxia/software/software.htm, and converted to amino acid sequences which were then aligned using the ClustalW [46] option. In order to maintain codon integrity the nucleotide sequences were then aligned to the amino acid alignment. Phylogenetic analyses were conducted using MEGA version 2.1 [47] – http://www.megasoftware.net. A maximum parsimony tree with branch confidence values based on 500 bootstrap replicates was constructed and the tree drawn in TreeViewPPC version 1.6.6 – http://taxonomy.zoology.gla.ac.uk/rod/treeview.html. Branches with less than 40% support were collapsed.
References
Burns ME, Baylor DA: Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu Rev Neurosci. 2001, 24: 779-805. 10.1146/annurev.neuro.24.1.779.
Ebrey T, Koutalos Y: Vertebrate photoreceptors. Prog Retin Eye Res. 2001, 20: 49-94. 10.1016/S1350-9462(00)00014-8.
Lythgoe JN: The Ecology of Vision. Oxford: Clarendon Press. 1979
Okano T, Kojima D, Fukada Y, Shichida Y, Yoshizawa T: Primary structures of chicken cone visual pigments: vertebrate rhodopsins have evolved out of cone visual pigments. Proc Natl Acad Sci U S A. 1992, 89: 5932-5936.
Okano T, Yoshizawa T, Fukada Y: Pinopsin is a chicken pineal photoreceptive molecule. Nature. 1994, 372: 94-97. 10.1038/372094a0.
Soni BG, Philp AR, Foster RG, Knox BE: Novel retinal photoreceptors. Nature. 1998, 394: 27-28. 10.1038/27794.
Chen P, Hao W, Rife L, Wang XP, Shen D, Chen J, Ogden T, Van Boemel GB, Wu L, Yang M, Fong HKW: A photic visual cycle of rhodopsin regeneration is dependent on Rgr . Nat Genet. 2001, 28: 256-260. 10.1038/90089.
Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J: Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 1997, 94: 9893-9898. 10.1073/pnas.94.18.9893.
Blackshaw S, Snyder SH: Parapinopsin, a novel catfish opsin localized to the parapineal organ, defines a new gene family. J Neurosci. 1997, 17: 8083-8092.
Blackshaw S, Snyder SH: Encephalopsin: a novel mammalian extraretinal opsin discretely localized in the brain. J Neurosci. 1999, 19: 3681-3690.
Hattar S, Liao H-W, Takao M, Berson DM, Yau K-W: Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002, 295: 1065-1070. 10.1126/science.1069609.
Bellingham J, Foster RG: Opsins and mammalian photoentrainment. Cell Tissue Res. 2002, 309: 57-71. 10.1007/s00441-002-0573-4.
Nathans J, Hogness DS: Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983, 34: 807-814.
Zhang H, Yokoyama S: Molecular evolution of the rhodopsin gene of marine lamprey, Petromyzon marinus . Gene. 1997, 191: 1-6. 10.1016/S0378-1119(96)00864-5.
Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM: The rhodopsin-encoding gene of bony fish lacks introns. Gene. 1995, 164: 273-277. 10.1016/0378-1119(95)00458-I.
Hunt DM, Wilkie SE, Bowmaker JK, Poopalasundaram S: Vision in the ultraviolet. Cell Mol Life Sci. 2001, 58: 1583-1598.
Nathans J, Thomas D, Hogness DS: Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986, 232: 193-202.
Yokoyama S, Yokoyama R: Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annu Rev Ecol Syst. 1996, 27: 543-567. 10.1146/annurev.ecolsys.27.1.543.
Goldsmith TH: Optimization, constraint, and history in the evolution of eyes. Q Rev Biol. 1990, 65: 281-322.
Jacobs GH: The distribution and nature of colour vision among the mammals. Biol Rev Camb Philos Soc. 1993, 68: 413-471.
Max M, McKinnon PJ, Seidenman KJ, Barrett RK, Applebury ML, Takahashi JS, Margolskee RF: Pineal opsin: a nonvisual opsin expressed in chick pineal. Science. 1995, 267: 1502-1506.
Moutsaki P, Bellingham J, Soni BG, David-Gray ZK, Foster RG: Sequence, genomic structure and tissue expression of carp (Cyprinus carpio L.) vertebrate ancient (VA) opsin. FEBS Lett. 2000, 473: 316-222. 10.1016/S0014-5793(00)01550-7.
Blackshaw S, Snyder SH: Developmental expression pattern of phototransduction components in mammalian pineal implies a light-sensing function. J Neurosci. 1997, 17: 8074-8082.
Shen D, Jiang M, Hao W, Tao L, Salazar M, Fong HKW: A human opsin-related gene that encodes a retinaldehyde-binding protein. Biochemistry. 1994, 33: 13117-13125.
Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD: A novel human opsin in the inner retina. J Neurosci. 2000, 20: 600-605.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Mount SM: Genomic sequence, splicing, and gene annotation. Am J Hum Genet. 2000, 67: 788-792. 10.1086/303098.
Shiang R, Murray JC, Morton CC, Buetow KH, Wasmuth JJ, Olney AH, Sanger WG, Goldberger G: Mapping of the human complement factor I gene to 4q25. Genomics. 1989, 4: 82-86.
Hameed A, Khaliq S, Ismail M, Anwar K, Mehdi SQ, Bessant D, Payne AM, Bhattacharya SS: A new locus for autosomal recessive RP (RP29) mapping to chromosome 4q32-q34 in a Pakistani family. Invest Ophthalmol Vis Sci. 2001, 42: 1436-1438.
Yokoyama S, Zhang H: Cloning and characterization of the pineal gland-specific opsin gene of marine lamprey (Petromyzon marinus). Gene. 1997, 202: 89-93. 10.1016/S0378-1119(97)00458-7.
Kusakabe T, Kusakabe R, Kawakami I, Satou Y, Satoh N, Tsuda M: Ci-opsin1, a vertebrate-type opsin gene, expressed in the larval ocellus of the ascidian Ciona intestinalis. FEBS Lett. 2001, 506: 69-72. 10.1016/S0014-5793(01)02877-0.
Stoltzfus A, Logsdon JM, Palmer JD, Doolittle WF: Intron "sliding" and the diversity of intron positions. Proc Natl Acad Sci U S A. 1997, 94: 10739-10744. 10.1073/pnas.94.20.10739.
Bellingham J, Wilkie SE, Morris AG, Bowmaker JK, Hunt DM: Characterisation of the ultraviolet-sensitive opsin gene in the honey bee, Apis mellifera . Eur J Biochem. 1997, 243: 775-781.
Rogozin IB, Lyons-Weiler J, Koonin EV: Intron sliding in conserved gene families. Trends Genet. 2000, 16: 430-432. 10.1016/S0168-9525(00)02096-5.
Gotoh O: Divergent structures of Caenorhabditis elegans cytochrome P450 genes suggest the frequent loss and gain of introns during the evolution of nematodes. Mol Biol Evol. 1998, 15: 1447-1459.
Rzhetsky A, Ayala FJ, Hsu LC, Chang C, Yoshida A: Exon/intron structure of aldehyde dehydrogenase genes supports the "introns-late " theory. Proc Natl Acad Sci U S A. 1997, 94: 6820-6825. 10.1073/pnas.94.13.6820.
Bellingham J, Whitmore D, Philp AR, Wells DJ, Foster RG: Zebrafish melanopsin: isolation, tissue localisation and phylogenetic position. Molecular Brain Research. 2002, 107: 128-136. 10.1016/S0169-328X(02)00454-0.
Hao W, Fong HKW: Blue and ultraviolet light-absorbing opsin from the retinal pigment epithelium. Biochemistry. 1996, 35: 6251-6256. 10.1021/bi952420k.
Hao W, Fong HKW: The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. J Biol Chem. 1999, 274: 6085-6090. 10.1074/jbc.274.10.6085.
Koyanagi M, Terakita A, Kubokawa K, Shichida Y: Amphioxus homologs of Go-coupled rhodopsin and peropsin having 11-cis – and all-trans-retinals as their chromophores. FEBS Lett. 2002, 531: 525-528. 10.1016/S0014-5793(02)03616-5.
Tarttelin EE, Bellingham J, Bibb LC, Foster RG, Hankins MW, Gregory-Evans K, Gregory-Evans CY, Wells DJ, Lucas RJ: Expression of opsin genes early in ocular development of humans and mice. Exp Eye Res. 2003, 76: 393-396. 10.1016/S0014-4835(02)00300-7.
Bibb LC, Holt JK, Tarttelin EE, Hodges MD, Gregory-Evans K, Rutherford A, Lucas RJ, Sowden JC, Gregory-Evans CY: Temporal and spatial expression patterns of the CRX transcription factor and its downstream targets. Critical differences during human and mouse eye development. Hum Mol Genet. 2001, 10: 1571-1579. 10.1093/hmg/10.15.1571.
Maden M: Retinoid signalling in the development of the central nervous system. Nat Rev Neurosci. 2002, 3: 843-853. 10.1038/nrn963.
Tatusova TA, Madden TL: BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999, 174: 247-250. 10.1016/S0378-1097(99)00149-4.
Xia X, Xie Z: DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001, 92: 371-373. 10.1093/jhered/92.4.371.
Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22: 4673-4680.
Kumar S, Tamura K, Jakobsen IB, Nei M: MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001, 17: 1244-1245. 10.1093/bioinformatics/17.12.1244.
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M: Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000, 289: 739-745. 10.1126/science.289.5480.739.
Acknowledgements
This work was supported by The Hammersmith Hospitals Trust Special Trustees (JB and DJW) and BBSRC (RGF).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
JB undertook the in silico and phylogenetic analysis, and participated in the preparation of the manuscript. DJW and RGF participated in the preparation of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Bellingham, J., Wells, D.J. & Foster, R.G. In silico characterisation and chromosomal localisation of human RRH (peropsin) – implications for opsin evolution. BMC Genomics 4, 3 (2003). https://doi.org/10.1186/1471-2164-4-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-4-3