Abstract
Background
Anthocyanins are a group of flavonoid compounds. As a group of important secondary metabolites, they perform several key biological functions in plants. Anthocyanins also play beneficial health roles as potentially protective factors against cancer and heart disease. To elucidate the anthocyanin biosynthetic pathway in Brassica rapa, we conducted comparative genomic analyses between Arabidopsis thaliana and B. rapa on a genome-wide level.
Results
In total, we identified 73 genes in B. rapa as orthologs of 41 anthocyanin biosynthetic genes in A. thaliana. In B. rapa, the anthocyanin biosynthetic genes (ABGs) have expanded and most genes exist in more than one copy. The anthocyanin biosynthetic structural genes have expanded through whole genome and tandem duplication in B. rapa. More structural genes located upstream of the anthocyanin biosynthetic pathway have been retained than downstream. More negative regulatory genes are retained in the anthocyanin biosynthesis regulatory system of B. rapa.
Conclusions
These results will promote an understanding of the genetic mechanism of anthocyanin biosynthesis, as well as help the improvement of the nutritional quality of B. rapa through the breeding of high anthocyanin content varieties.
Similar content being viewed by others
Background
Flavonoids comprise a major group of secondary metabolites, which exhibit a wide range of biological functions in plants [1, 2]. Anthocyanin pigments and flavonol co-pigments are the two major flavonoid compounds, which serve as attractants of pollinators and seed dispersers. They also play an important role in protecting plants against abiotic and biotic stresses [3]. Anthocyanins, like other flavonoid compounds, are known as potent antioxidants [4]. The beneficial health roles of anthocyanins have received considerable attention as they are potentially protective factors against cancer and heart disease [5]. Therefore, a comprehensive understanding of anthocyanin biosynthesis is important for developing foods that are rich in anthocyanins to meet the increasing demand for health-promoting components in our daily diet.
The biosynthetic pathways of anthocyanins have been well characterized [6] and the corresponding genes have been isolated from various plants. In the model plant A. thaliana, the biosynthesis, regulation and transport of anthocyanins, specifically most of the structural genes and regulatory proteins involved in anthocyanin synthesis, have been identified and functionally characterized in the last two decades [7–9]. These researches have played important roles in the comprehensive understanding of anthocyanin biosynthesis and revealed the accumulation and metabolic profiles of anthocyanins in A. thaliana.
Brassica rapa comprises a variety of vegetables, among which Chinese cabbage (Brassica rapa L. ssp. pekinensis) and pakchoi (Brassica rapa L. ssp. chinensis) are the two most consumed vegetables in China and throughout East Asia. B. rapa vegetables provide dietary fiber, vitamin C, and anti-cancer glucosinolates [10], and are also a potentially important source of dietary flavonols [11]. Several varieties of B. rapa are red or purple, such as some kinds of pakchoi, turnip (Brassica rapa ssp. rapa) and Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). The purple pigments in B. rapa have been identified as anthocyanins [12]. Nevertheless, the genetic mechanism of this purple phenotype is unclear. Because of the previous absence of genome information, little is known about the genes involved in the anthocyanin biosynthetic pathways in B. rapa[13, 14]. A complete understanding of the structural and regulatory genes involved is very important for elaborating the mechanism of anthocyanin biosynthesis in B. rapa, as well as for the breeding of new B. rapa varieties rich in anthocyanins. With the reference genome and gene annotation information for B. rapa ‘Chiifu’ [15], we now have the chance to systematically study the anthocyanin biosynthetic genes (ABGs) in B. rapa.
Both B. rapa and A. thaliana belong to the cruciferae family. B. rapa (A genome) has undergone whole genome triplication since its divergence from A. thaliana, followed by extensive gene loss [15, 16]. In B. rapa, the level of gene loss among the three subgenomes is unequal, with one subgenome, the Least Fractionated (LF) subgenome, having retained roughly 70% of its genes during fractionation after triplication, while the other two subgenomes, termed the Medium and Most Fractionated (MF1 and MF2, respectively) subgenomes, have retained much fewer genes [15, 16].
Whole genome duplication (WGD) is a major way of gene copy number expansion in plants. As reported previously, gene families expanded by WGD can maintain the proper balance in biological networks or cascades [17]. After WGD and subsequent gene fractionation, the number of genes that respond to abiotic and biotic stresses or with membrane protein functions tends to be increased [18]. In B. rapa, the genes expanded through whole genome triplication (WGT) tend to come from functional categories such as transcriptional regulation, ribosomes, response to abiotic or biotic stimuli, response to hormonal stimuli, cell organization, and transporter functions [15]. However, no detailed information about the status of ABGs after the WGT in B. rapa has been available till now. To obtain comprehensive information on the anthocyanin biosynthetic pathway in B. rapa and look into the effect of WGT on these genes, comparative genomic analysis between B. rapa and A. thaliana was performed here based on the reference genome of B. rapa[15]. We present several interesting observations about the evolutionary history of ABGs in B. rapa, the similarities between the anthocyanin biosynthetic pathways in A. thaliana and B. rapa, the synteny of ABGs between A. thaliana and B. rapa, and the expansion and retention of ABGs in B. rapa. The results of our systematic analysis of the complete set of ABGs in B. rapa will promote the understanding of the genetic mechanism of anthocyanin biosynthesis and the anthocyanin profiles/accumulation in B. rapa crops.
Results and discussion
ABGs in B. rapaidentified by comparative genomic analysis
The ABGs have expanded in the genome of B. rapa. In A. thaliana, 41 ABGs have been reported, including 24 structural genes that encode anthocyanin biosynthesis enzymes, 16 regulatory genes encoding transcriptional factors, and one transport gene that is required for anthocyanin transportation (Table 1; Additional file 1: Table S1). Based on a combination of syntenic and non-syntenic homology analysis, 73 B. rapa anthocyanin biosynthetic genes (BrABGs) were identified, representing homologs of 39 out of the 41 AtABGs. The other two AtABGs were not found in B. rapa.
Among the 73 BrABGs, 67 were syntenic orthologs of 38 AtABGs; only 8.2% of BrABGs had no syntenic relationship. Multiple copies of BrABGs in B. rapa syntenic to genes in A. thaliana were generated from the WGT, whereas 28 of the 38 AtABGs had less than three syntenic orthologs as a result of gene fractionation following triplication as described above. Furthermore, the amino acid sequence identities for pairs of syntenic genes (80.35%) and non-syntenic genes (80.43%) did not show a statistical difference. The BrABGs of different functions in the anthocyanin biosynthesis pathway had different sequence identities to their counterparts in A. thaliana (Additional file 1: Table S1). The homologous pairs of structural genes encoding anthocyanin biosynthesis enzymes shared significantly higher amino acid identity (82.10%) than those encoding transcriptional factors (76.79%), which regulate the expression of structural genes (P < 0.05), indicating that the structural genes were more highly conserved than regulatory genes. It is reasonable that excessive mutation of structural genes would likely block the biosynthesis of anthocyanins, reducing the fitness of the plant.
Of the 73 BrABGs, 72 were mapped to the 10 chromosomes of B. rapa, with three, 12, 12, seven, 10, four, six, one, 12, and five BrABGs located on chromosomes A01-A10 of B. rapa genome V1.5, respectively (Figure 1). The remaining gene, Bra035004, an ortholog of UGT79B1, was anchored on Scaffold000100, which has not yet been mapped onto a chromosome. Whole genome analysis established that the three subgenomes in B. rapa could be distinguished by the degree of gene density [15, 16]. With this subgenome information, we then assigned all BrABGs to the three subgenomes: LF, MF1 and MF2. In total, 31, 20 and 21 genes were located on LF, MF1 and MF2, respectively. There were more genes located on LF, while almost equal numbers of genes were distributed on MF1 and MF2. Of the 67 syntenic orthologs, 30 were on LF, 17 were on MF1 and 20 were on MF2; the number of syntenic orthologs distributed on LF was a little less than the sum of those on MF1 and MF2. These results show that the distribution of BrABGs is consistent with the gene fractionation status at the whole genome level [15, 16]. Based on the determination of these BrABGs, the anthocyanin biosynthetic pathway in B. rapa was thus established.
ABGs are over-retained in B. rapa
Compared with A. thaliana, the B. rapa genome has undergone a whole genome triplication [15]. Most of the ABGs were present in multiple copies in B. rapa. In our study, the 73 BrABGs represent 0.18% of the 41,174 predicted genes in B. rapa; accordingly, there are 41 AtABGs genes representing 0.15% of all A. thaliana genes. With the total numbers of A. thaliana and B. rapa genes as a background, we found that the expansion levels of structural, regulatory, and transport genes and the total number of BrABGs were similar to the whole-genome gene expansion level in B. rapa (P > 0.05, Table 2). We also analyzed the number and ratio of single copy to multiple copy paralogous genes to reveal the retention status of the BrABGs after the whole genome triplication. We used the ratio of total number of single to multiple copy (two and three) paralogous genes (genes in the same tandem array were only counted once) of the whole B. rapa genome as a background (1.77; 15795/8935; [15]) and found that the total BrABGs (0.74; 14/19) and regulatory genes (0.20; 2/10) were significantly lower than that of the background, indicating over-retention of total and regulatory ABGs in B. rapa. The structural and transport genes showed no significant difference to the background (Table 3).
The anthocyanin biosynthetic structural genes have expanded through whole genome and tandem duplication in B. rapa
Gene copy numbers can be expanded in four major ways: WGD, tandem duplication (TD), segmental duplication, and gene transposition duplication [19]. We found that the structural BrABGs were expanded through both WGD and TD. We identified 46 BrABG structural genes as homologs of 24 AtABG structural genes. The almost-doubled number of homologs indicated that anthocyanin biosynthetic structural genes were expanded in B. rapa. Of the 46 structural genes, 41 were located at 33 loci that had syntenic relationships to 21 AtABG structural gene loci. Among the 41 syntenic orthologs, 14 BrABGs came from six tandem arrays, which accounted for about 30% of all structural genes in B. rapa. These data showed that both TD and WGT contributed to the expansion of anthocyanin biosynthetic structural genes in B. rapa. The 46 genes were distributed in different subgenomes of B. rapa, with 19, 12 and 14 genes located on LF, MF1 and MF2, respectively, and one gene not anchored on any chromosome in the current version of the B. rapa genome.
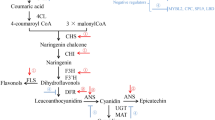
Anthocyanins are derived from branches of the flavonoid pathway, which starts with phenylalanine via the general phenylpropanoid pathway. The phenylpropanoid pathway contains three major genes: PAL, C4H and 4CL. Two types of correlated structural genes can be distinguished in the flavonoid biosynthetic pathway: Early Biosynthetic Genes (EBGs) and Late Biosynthetic Genes (LBGs) [20]. The EBGs, which include CHS, CHI, F3H, F3’H, and FLS, lead to the production of flavonols and other flavonoid compounds, while the LBGs, which include DFR, ANS/LDOX, and UFGT, lead to the production of anthocyanins [9]. The phenylpropanoid pathway genes and EBGs are upstream genes while the LBGs are downstream genes in the anthocyanin biosynthetic pathway. The downstream genes are specifically for anthocyanin biosynthesis [21].
The upstream structural genes have been expanded not only by WGD, but also TD. There are 21 homologs of the nine A. thaliana phenylpropanoid pathway genes in B. rapa (Table 1). AtC4H has five syntenic orthologs in B. rapa. BrC4H2 and BrC4H3, BrC4H4 and BrC4H5 are from two tandem arrays located in MF1 and MF2, respectively, while BrC4H1 is in LF (Table 1). This shows that BrC4Hs have expanded by both WGD and TD. At4CL2 and At4CL5 have not expanded by whole genome duplication, but their homologs in B. rapa form two tandem arrays, with one array containing four genes and the other containing two genes (Table 1).
The biosynthesis of flavonol glycosides and anthocyanins shares common substrates: dihydroflavonols [22]. The synthesis of flavonol aglycones has long been attributed to a single enzyme, flavonol synthase (FLS), which competes with several other enzymes for dihydroflavonol substrates [23]. There are six FLS genes in A. thaliana: AtFLS1 to 6. These six genes are all located on chromosome 5, with AtFLS2-5 arranged in a 7.5-Kb tandem array [21]. There are also six FLS genes in B. rapa: BrFLS1, BrFLS2, BrFLS3.1, BrFLS3.2, BrFLS3.3, and BrFLS4 (Table 1). The tandem array AtFLS2-5 has syntenic loci in all the three subgenomes of B. rapa: BrFLS2 and BrFLS3.1 in LF, BrLFS3.2 and BrFLS3.3 in MF1, and BrFLS4 in MF2 (Figure 2). The tandem array was duplicated by WGD, but only some of the four tandemly duplicated genes have been retained, with two, two, and one in LF, MF1 and MF2, respectively (Figure 2). No homologous genes for AtFLS6 were found in B. rapa, neither syntenic nor non-syntenic orthologs. A phylogenetic tree was constructed using the amino acid sequences of the six AtFLS genes, the six homologs in B. rapa and a FLS gene of Vitis vinifera to reveal the relationship between these genes (Figure 3). In the phylogenetic tree, AtFLS1 and BrFLS1, AtFLS2 and BrFLS2, AtFLS3 and BrFLS3.1, 3.2, and 3.3, and AtFLS4 and BrFLS4 were clustered together. These results confirmed the hypothesis proposed by Owens et al. (2008) that the duplications leading to the amplification of the FLS gene family were ancient events [21], from at least before the divergence of B. rapa and A. thaliana.
Arrangement of the AtFLSs in A. thaliana genome as well as their corresponding syntenic homologs in B. rapa chromosomes and subgenomes showed duplication, retention and distribution of BrFLSs. All six AtFLSs are located on chromosome 5. AtFLS2 to −5 are clustered in a 7.5-kb region. AtFLS1 only has one syntenic ortholog BrFLS1 on chromosome A10 and subgenome LF. There are no orthologs of AtFLS6. AtFLS2 to −5 have five synteny orthologs distributed in different chromosomes and subgenomes. The black and green bars represent the chromosomes of A. thaliana and B. rapa. The LF, MF1, and MF2 in brackets below gene names indicate the different subgenomes of B. rapa.
DFR, LDOX/ANS and UFGTs are the major LBGs (downstream structural genes) of the anthocyanin biosynthetic pathway. DFR (dihydroflavonol-4-reductase) catalyzes the first committed reaction to generate anthocyanins. In A. thaliana, the gene AtDFR is known to encode a functional DFR [24]. DFR was not expanded in B. rapa; only one gene, BrDFR, was identified as an ortholog of AtDFR. Leucoanthocyanidin dioxygenase/anthocyanidin synthase (LDOX/ANS) catalyzes the formation of anthocyanidin, the first colored compound in the anthocyanin biosynthetic pathway [25]. Two syntenic orthologs in B. rapa, BrANS1 and BrANS2, were identified by comparative genomic analysis with A. thaliana.
The gene copy number ratios between B. rapa and A. thaliana were 2.3 (21:9), 2 (18:9) and 1.4 (7:5) for the phenylpropanoid pathway genes, EBGs and LBGs, respectively. In addition to WGT, some phenylpropanoid pathway genes and EBGs (which are the upstream structural genes) were also expanded by TD (Table 1). These results show that more upstream structural genes were retained than downstream structural genes of the anthocyanin biosynthetic pathway. The redundancy of the upstream genes may guarantee products for successful downstream anthocyanin synthesis.
More negative regulatory genes are retained in the anthocyanin biosynthesis regulatory system of B. rapa
Anthocyanin biosynthesis is regulated mostly by the coordinated transcriptional control of the structural genes. The transcriptional control of ABG structural genes has been intensively studied [7]. In A. thaliana, anthocyanin biosynthetic regulatory genes can be divided into two groups: positive and negative regulatory genes. For positive regulation of anthocyanin biosynthesis in A. thaliana, the spatial and temporal expression of structural genes is mainly determined by R2R3-MYB, basic helix-loop-helix (bHLH) and WD40-type transcriptional factors and their interaction [26]. Four R2R3-MYB (PAP1, PAP2, MYB113 and MYB114) transcription factors and three bHLH (TT8, GL3 and EGL3) proteins combine with the WD40 repeat protein (TTG1) to form ternary transcriptional complexes that activate several anthocyanin biosynthetic structural genes, especially in the later steps (LBGs) of the flavonoid pathway [27–30]. Other R2R3-MYB proteins (MYB11, MYB12, and MYB111) regulate the structural genes of the anthocyanin biosynthetic pathway independently, especially the early steps (EBGs) of the flavonoid pathway [31, 32]. Two single-repeat R3-MYB transcription factors, MYBL2 and CPC (CAPRICE), and three members of the LATERAL ORGAN BOUNDARY DOMAIN (LBD) gene family, LBD37, LBD38, and LBD39, are negative regulators of anthocyanin biosynthesis in A. thaliana[33–36]. The regulatory genes in anthocyanin biosynthetic pathway of B. rapa were identified as well as the duplication and retention of these genes were analyzed. Detailed information on these biosynthetic regulatory genes in B. rapa is presented below.
MYB11, MYB12, and MYB111 activate EBGs of the anthocyanin biosynthetic pathway in A. thaliana[37]. In B. rapa, there were two syntenic orthologs of AtMYB12 and three syntenic orthologs of AtMYB111 (Table 1), but no homologs of AtMYB11 were found. These results show that the homolog of AtMYB11 was lost, while the homologs of AtMYB12 and AtMYB111 were over-retained after the WGT of B. rapa.
AtPAP1, AtPAP2, AtMYB113, and AtMYB114 (the four R2R3-MYB transcription factors) activate the LBGs by forming ternary complexes with bHLH and WD40 proteins. These four genes are all distributed on chromosome 1 of A. thaliana. AtPAP2, AtMYB113 and AtMYB114 are present as a three-gene tandem array. The four AtABGs have three orthologs in B. rapa: Bra004162, Bra039763 and Bra001917. The high sequence similarity and tandem relationship of these At R2R3-MYB genes made it difficult to name the three Br genes by synteny or homology analysis. Bra004162 and Bra037763 are two syntenic orthologs of the three-gene tandem array. Tandem arrays were not detected in B. rapa, indicating that the tandem duplication may have occurred in A. thaliana after its divergence from B. rapa, or that the redundant tandem arrays were lost through the influence of WGT in B. rapa[38]. These data suggest that genes encoding R2R3-MYBs, which form MBW complexes and regulate anthocyanin biosynthesis, were lost after the WGT in B. rapa compared with those in A. thaliana. Bra001917 is a non-syntenic ortholog of AtPAP1, AtPAP2, AtMYB113, and AtMYB114, but its syntenic gene in A. thaliana is AT3G23250, which encodes AtMYB15. A phylogenetic tree was constructed by aligning the amino acid sequences of AtPAP1, AtPAP2, AtMYB113, AtMTB114, Bra001917, Bra004162, Bra039763 and Zmp1 (a gene encoding a R2R3-MYB transcriptional factor in maize as the outgroup, Figure 4). In the phylogenetic tree, the four A. thaliana genes and three B. rapa genes are clustered together in one branch, while AtMYB15 and Zmp1 form another branch.
Another important group of transcriptional factors is the basic helix-loop-helix (bHLH) gene family, which regulates anthocyanin biosynthesis through formation of MBW ternary complexes. In A. thaliana, the bHLH transcriptional factors TT8 (TRANSPARENT TESTA 8), GL3 (GLABROUS 3) and EGL3 (ENHANCER OF GLABRA 3) interact with TTG1 (WD40 protein) and PAP1, PAP2, MYB113, or MYB114 to form multiple MBW complexes that then activate LBGs. In B. rapa, there are four BrbHLH genes, BrTT8, BrGL3, BrEGL3.1 and BrEGL3.2, corresponding to three bHLH genes in A. thaliana. The BrbHLH genes in B. rapa seem to have undergone relatively extensive fractionation after WGT.
TTG1 (Transparent Testa Glabra 1) is the only gene that encodes a WD40 protein involved in the regulation of anthocyanin biosynthesis in A. thaliana[39]. In B. rapa, there are two paralogous genes, BrTTG1.1 and BrTTG1.2, which are syntenic orthologs of AtGTT1. The amino acid sequence of BrTTG1.2 is shorter than those of BrTTG1.1 and AtTTG1. BrTTG1.2 has lost its first WD40 repeat domain, N-terminal and C-terminal regions (Figure 5). A previous study showed that the C-terminal region was vital for the structural and function of TTG1 [40]. A mutant of A. thaliana lacking 25 amino acid residues at the C-terminus showed a severe phenotype with no anthocyanins in the testa [40]. These results suggest that BrTTG1.2 might be non-functional because of large sequence deletions and has become a pseudogene after WGT.
There are several transcription factors including two R3-type single MYB proteins, MYBL2 and CPC, and three N/NO3− induced members of the LBD gene family that act as negative regulators of anthocyanin biosynthesis in A. thaliana. AtMYBL2 is a transcriptional repressor that negatively regulates anthocyanin biosynthesis by interacting with TT8 and MBW complexes [33, 34]. In B. rapa, there are two syntenic orthologs, BrMYBL2.1 and BrMYBL2.2, found in LF and MF1, respectively. AtCPC, another single repeat R3-MYB transcription factor, works as a negative regulator of anthocyanin biosynthesis [35]. The AtCPC gene has two syntenic orthologs in B. rapa, BrCPC1 and BrCPC2, which both share more than 90% amino acid sequence identity with their At ortholog. LBD37, LBD38 and LBD39 negatively regulate the late anthocyanin-specific steps by repressing PAP1 and PAP2 under N/NO3− induction [36]. There are seven syntenic orthologs of AtLBD37, AtLBD38 and AtLBD39 in B. rapa (Table 1).
The regulatory genes have expanded mainly through WGT in B. rapa. In total, 25 BrABG regulatory genes were identified as homologs of 16 AtABG regulatory genes. Fourteen BrABG positive regulatory genes were identified as homologs of 11 AtABGs, while 11 BrABG negative regulatory genes were identified as homologs of 5 AtABGs. The copy numbers of the positive regulatory genes did not show a significant change between B. rapa and A. thaliana, but the number of negative regulatory genes was almost doubled in B. rapa. Comparing the copy numbers of the regulatory genes, we found that more negative than positive regulatory genes were retained in the B. rapa genome.
Anthocyanins play important roles in responding to abiotic and biotic stresses for plants. B. rapa comprises a variety of vegetables with rich morphological diversity. Several varieties accumulate variant kinds and contents of anthocyanins in different tissues, while more varieties of B. rapa present green with no anthocyanins accumulated (data not shown in this paper). So the copy numbers of negative and positive regulatory genes will help us to understand the metabolic characteristics of anthocyanin in B. rapa.
Conclusions
Anthocyanin biosynthetic genes (ABGs) were identified based on whole-genome comparative analysis between A. thaliana and B. rapa. Anthocyanin biosynthetic pathways including 73 genes were established in B. rapa. Multiple copies of the BrABGs were generated by WGD and retained synteny with their orthologs in A. thaliana, but most genes appeared to comprise less than three copies because of gene loss following WGT. More upstream structural genes of the anthocyanin biosynthetic pathway have been retained than downstream. Based on the presence of these homologous structural genes, the anthocyanin biosynthetic pathway in B. rapa was then established. More negative regulatory genes have been retained than positive by comparing the copy numbers. The composition of BrABGs could help us to explain the metabolic profiles of anthocyanin accumulation and elaborate the genetic mechanism of anthocyanin biosynthesis in B. rapa.
There has been little research on anthocyanin biosynthetic genes in B. rapa. Here, we identified anthocyanin biosynthetic genes systematically at the whole genome level. The determination of a complete set of anthocyanin biosynthetic genes in B. rapa provides a valuable resource for the study of anthocyanin-related traits and genetic improvement of the anthocyanin nutritional quality of B. rapa.
Methods
Database for ABGs identification in B. rapa
The complete sets of gene sequences in A. thaliana involved in the anthocyanin biosynthetic pathway were downloaded from the TAIR database (http://www.arabidopsis.org/). The B. rapa genome sequence (version 1.5) and gene sequences from BRAD (http://brassicadb.org/brad/) [41] were used to identify the ABGs in B. rapa.
Identification of homologous genes between B. rapa and A. thaliana
We used the anthocyanin biosynthetic gene and protein sequences of A. thaliana to align with the genome and protein sequences of B. rapa using BLASTN and BLASTP with a cut off E-value ≤ 1E−10 and coverage ≥ 0.75. We identified syntenic orthologs between A. thaliana and B. rapa from BRAD (http://brassicadb.org/brad/), which were determined by both sequence similarity (cutoff: E ≤ 10−20 ) and the collinearity of flanking genes [42].
Phylogenetic analysis of gene sequences
A phylogenetic tree was constructed by the neighbor-joining method using MEGA4 [43]. The stability of tree nodes was tested by bootstrap analysis with 1000 iterations.
Availability of supporting data
All the anthocyanin biosynthetic genes of A. thaliana referred in this paper were retrieved from the TAIR database (http://www.arabidopsis.org/).
The B. rapa genome sequence (version 1.5), as well as CDS and protein sequences of BrABGs were retrieved from the Brassica database (BRAD) (http://brassicadb.org/brad/).
The protein sequences of VvFLS (BAE75808) and ZmP1 (P27898) used as outgroups to construct the phylogenetic trees in this paper were downloaded from GeneBank (NCBI) (http://www.ncbi.nlm.nih.gov/).
Abbreviations
- LF:
-
Least fractionated
- MF1:
-
Medium fractionated
- MF2:
-
Most fractionated
- WGD:
-
Whole genome duplication
- WGT:
-
Whole genome triplication
- ABGs:
-
Anthocyanin biosynthetic genes
- TD:
-
Tandem duplication
- EBGs:
-
Early biosynthetic genes
- LBGs:
-
Late biosynthetic genes
- bHLH:
-
basic helix-loop-helix
- MBW:
-
MYB-bHLH-WD40
- PAL:
-
Phenylalanine ammonia-lyase
- C4H:
-
Cinnamate-4-hydroxylase
- 4CL:
-
4-coumarate:CoA ligase
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- F3H:
-
Flavanone 3-hydroxylase
- F3’H:
-
Flavonoid 3’-hydroxylase
- FLS:
-
Flavonol synthase
- DFR:
-
Dihydroflavonol-4-reductase
- LDOX:
-
Leucoanthocyanidin dioxygenase
- ANS:
-
Anthocyanidin synthase
- UGT79B1:
-
Anthocyanin 3-O-glucoside: 2 -O-xylosyltransferase
- UGT75C1:
-
Anthocyanin 5-O-glucosyltransferase
- UGT78D2:
-
Anthocyanidin 3-O-glucosyltransferase
- PAP1/2:
-
Production of anthocyanin pigment 1/2
- TT8/19:
-
Transparent testa 8/19
- GL3:
-
Glabrous 3
- EGL3:
-
Enhancer of glabrous 3
- TTG1:
-
Transparent testa glabrous 1
- MYBL2:
-
MYB-like 2
- CPC:
-
CAPRICE
- LBD:
-
Lateral organ boundary domain.
References
Harborne JB, Baxter H, Moss GP: Phytochemical Dictionary: A Handbook of Bioactive Compounds from Plants. 1999, London: Boca Raton: CRC Press, Taylor & Francis, 2
Andersen OM, Markham KR: Flavonoids: Chemistry, Biochemistry and Applications. 2006, London: Boca Raton: CRC Press, Taylor & Francis
Bradshaw H, Schemske DW: Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003, 426 (6963): 176-178. 10.1038/nature02106.
Yamasaki H, Sakihama Y, Ikehara N: Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. 1997, 115 (4): 1405-1412.
Steyn W, Wand S, Holcroft D, Jacobs G: Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol. 2002, 155 (3): 349-361. 10.1046/j.1469-8137.2002.00482.x.
Holton TA, Cornish EC: Genetics and biochemistry of anthocyanin biosynthesis. The Plant Cell. 1995, 7 (7): 1071-10.1105/tpc.7.7.1071.
Broun P: Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol. 2005, 8 (3): 272-279. 10.1016/j.pbi.2005.03.006.
Winkel-Shirley B: Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126 (2): 485-493. 10.1104/pp.126.2.485.
Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M: Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006, 57: 405-430. 10.1146/annurev.arplant.57.032905.105252.
Fahey JW, Talalay P, Gustine DI, Flores HE: The role of crucifers in cancer chemoprotection. Phytochemicals and Health. 1995, Rochville: American Society of Plant Physiologist, 87-93.
Rochfort SJ, Imsic M, Jones R, Trenerry VC, Tomkins B: Characterization of flavonol conjugates in immature leaves of pak choi [Brassica rapa L. ssp. chinensis L. (Hanelt.)] by HPLC-DAD and LC-MS/MS. J Agric Food Chem. 2006, 54 (13): 4855-4860. 10.1021/jf060154j.
Podsędek A: Natural antioxidants and antioxidant capacity of Brassica vegetables: A review. LWT-Food Science and Technology. 2007, 40 (1): 1-11. 10.1016/j.lwt.2005.07.023.
Kim C, Park S, Kikuchi S, Kwon S, Park S, Yoon U, Park D, Seol Y, Hahn J, Park S: Genetic analysis of gene expression for pigmentation in Chinese cabbage (Brassica rapa). BioChip Journal. 2010, 4 (2): 123-128. 10.1007/s13206-010-4206-9.
Burdzinski C, Wendell DL: Mapping the anthocyaninless (anl) locus in rapid-cycling Brassica rapa (RBr) to linkage group R9. BMC Genet. 2007, 8 (1): 64-10.1186/1471-2156-8-64.
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J-H, Bancroft I, Cheng F: The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011, 43 (10): 1035-1039. 10.1038/ng.919.
Cheng F, Wu J, Fang L, Sun S, Liu B, Lin K, Bonnema G, Wang X: Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012, 7 (5): e36442-10.1371/journal.pone.0036442.
Freeling M, Thomas BC: Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006, 16 (7): 805-814. 10.1101/gr.3681406.
Rizzon C, Ponger L, Gaut BS: Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006, 2 (9): e115-10.1371/journal.pcbi.0020115.
Sémon M, Wolfe KH: Consequences of genome duplication. Current opinion in genetics & development. 2007, 17 (6): 505-512. 10.1016/j.gde.2007.09.007.
Petroni K, Tonelli C: Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181 (3): 219-229. 10.1016/j.plantsci.2011.05.009.
Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BS: Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 2008, 147 (3): 1046-1061. 10.1104/pp.108.117457.
Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM: Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 2003, 131 (3): 259-268. 10.1023/A:1024018729349.
Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N: Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol J. 2007, 2 (10): 1214-1234. 10.1002/biot.200700084.
Shirley BW, Hanley S, Goodman HM: Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. The Plant Cell Online. 1992, 4 (3): 333-347. 10.1105/tpc.4.3.333.
Pelletier M, Murrell J, Shirley B: Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of "early" and "late" genes. Plant Physiol. 1997, 113 (4): 1437-1445. 10.1104/pp.113.4.1437.
Koes R, Verweij W, Quattrocchio F: Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10 (5): 236-242. 10.1016/j.tplants.2005.03.002.
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C: Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell Online. 2000, 12 (12): 2383-2393. 10.1105/tpc.12.12.2383.
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A: A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003, 130 (20): 4859-4869. 10.1242/dev.00681.
Tohge T, Nishiyama Y, Hirai M, Yano M, Nakajima J-i, Awazuhara M, Inoue E, Takahashi H, Goodenowe D, Kitayama M, Noji M, Yamazaki M, Saito K: Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. The Plant journal. 2005, 42 (2): 218-235. 10.1111/j.1365-313X.2005.02371.x.
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM: Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal. 2008, 53 (5): 814-827. 10.1111/j.1365-313X.2007.03373.x.
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B: Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal. 2007, 50 (4): 660-677. 10.1111/j.1365-313X.2007.03078.x.
Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B: Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111- independent flavonol glycoside accumulation. New Phytol. 2010, 188 (4): 985-1000. 10.1111/j.1469-8137.2010.03421.x.
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L: MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. The Plant Journal. 2008, 55 (6): 940-953. 10.1111/j.1365-313X.2008.03564.x.
Matsui K, Umemura Y, Ohme‒Takagi M: AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. The Plant Journal. 2008, 55 (6): 954-967. 10.1111/j.1365-313X.2008.03565.x.
Zhu H-F, Fitzsimmons K, Khandelwal A, Kranz RG: CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant. 2009, 2 (4): 790-802. 10.1093/mp/ssp030.
Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R: Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. The Plant Cell Online. 2009, 21 (11): 3567-3584. 10.1105/tpc.109.067041.
Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B: Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol. 2005, 57 (2): 155-171. 10.1007/s11103-004-6910-0.
Fang L, Cheng F, Wu J, Wang X: The impact of genome triplication on tandem gene evolution in Brassica rapa. Frontiers in plant science. 2012, 3:
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC: The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell Online. 1999, 11 (7): 1337-1349. 10.1105/tpc.11.7.1337.
Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW: The TTG1 Gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994, 166 (2): 740-754. 10.1006/dbio.1994.1352.
Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X: BRAD, the genetics and genomics database for Brassica plants. BMC plant biology. 2011, 11: 136-10.1186/1471-2229-11-136.
Cheng F, Wu J, Fang L, Wang X: Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Frontiers in plant science. 2012, 3:
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24 (8): 1596-1599. 10.1093/molbev/msm092.
Acknowledgements
The work was funded by the National High Technology R&D Program of China (2012AA100101), the National Program on Key Basic Research Projects of China (The 973 Program: 2012CB113900, 2013CB127000 and 2013CB127006) and International Joint Research Grant of Ministry of Science and Technology, P. R. China (2011DFR31180), as well as National Natural Science Foundation of China (NSFC grant: 31301771 and 31201628). Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P. R. China and the Sino-Dutch Joint Lab of Horticultural Genomics Technology in Beijing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NG searched and listed the ABGs from the references and database, performed the detailed analysis of BrABGs by comparative genomics, wrote and revised the manuscript. FC and BL identified the BrABGs by syntenic and non-syntenic homology analysis, respectively. SZ provided information about the AtABGs. XW, JW and FC conceived the idea and critically reviewed the manuscript. XW, JW and JL coordinated and designed the study. All the authors have read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Guo, N., Cheng, F., Wu, J. et al. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics 15, 426 (2014). https://doi.org/10.1186/1471-2164-15-426
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-15-426