Abstract
Background
microRNAs (miRNAs) are endogenous, noncoding, small RNAs that have essential regulatory functions in plant growth, development, and stress response processes. However, limited information is available about their functions in sexual reproduction of flowering plants. Pollen development is an important process in the life cycle of a flowering plant and is a major factor that affects the yield and quality of crop seeds.
Results
This study aims to identify miRNAs involved in pollen development. Two independent small RNA libraries were constructed from the flower buds of the male sterile line (Bcajh97-01A) and male fertile line (Bcajh97-01B) of Brassica campestris ssp. chinensis. The libraries were subjected to high-throughput sequencing by using the Illumina Solexa system. Eight novel miRNAs on the other arm of known pre-miRNAs, 54 new conserved miRNAs, and 8 novel miRNA members were identified. Twenty-five pairs of novel miRNA/miRNA* were found. Among all the identified miRNAs, 18 differentially expressed miRNAs with over two-fold change between flower buds of male sterile line (Bcajh97-01A) and male fertile line (Bcajh97-01B) were identified. qRT-PCR analysis revealed that most of the differentially expressed miRNAs were preferentially expressed in flower buds of the male fertile line (Bcajh97-01B). Degradome analysis showed that a total of 15 genes were predicted to be the targets of seven miRNAs.
Conclusions
Our findings provide an overview of potential miRNAs involved in pollen development and interactions between miRNAs and their corresponding targets, which may provide important clues on the function of miRNAs in pollen development.
Similar content being viewed by others
Background
Pollen, as the male gametophyte, participates in sexual reproduction in angiosperms and influences seed yield and quality. Pollen development is one of the most fascinating and critical processes that are critical for successful reproduction in the life cycle of flowering plants [1–3]. By employing a variety of resources and novel techniques, scientists have made significant progress in pollen research toward a deeper understanding of pollen development. In 2010, the phytohormone brassinosteroid was verified to control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development, such as SPL/NZZ, EMS1/EXS, DYT1, TDF1, AtMYB103, AMS, MS1, and MS2[4]. Honys and Twell conducted a genome-wide study on pollen transcriptome in Arabidopsis based on microarray analysis. Over 3500 genes were predicted to be expressed in pollen, out of which more than 1400 genes were pollen-specific [5]. A large-scale genetic screen was conducted in Arabidopsis, and a number of genes were identified to be involved in pollen exine production, including fatty acid ω-hydroxylase CYP704B1, putative glycosyl transferases At1g27600 and At1g33430, 4-coumarate-coenzyme A ligase 4CL3, and polygalacturonase QUARTET3[6]. Until now, a series of genes which were successively renamed Brassica campestris Male Fertile (BcMF) 2[7], BcMF3[8], BcMF4[9], up to BcMF21[10], have been demonstrated to be involved in pollen development in Brassica campestris. For example, among these genes, one type was polygalacturonase gene, which participated in pollen extine or intine development, such as BcMF6[11], BcMF2[7], and BcMF9[12]. These previous studies provide important information for understanding the gene regulatory networks of pollen development. However, the mechanisms that underlie pollen development remain unclear, and further studies must be conducted.
miRNAs are a newly identified class of endogenous non-coding small RNAs that have become a research hotspot because of their negative regulatory function in gene expression at the posttranscriptional level by degrading target mRNAs or repressing gene translation. Numerous studies have previously shown that miRNAs have important functions in regulating a wide range of plant developmental processes, including lateral root development [13], leaf development and polarity [14], vegetative phase change [15], flowering time and floral organ identity [16, 17], plant nutrient homeostasis [18], signal transmission [19], and response to environmental biotic and abiotic stresses [20, 21]. Among miRNAs that are known to function in a variety of plant development processes, few miRNAs in pollen tissue have been reported. Wei et al. identified 292 known miRNAs and 75 novel miRNAs in Oryza sativa. A total of 103 out of the 292 known miRNAs were enriched in developing pollen, and more than half of the 75 novel miRNAs displayed pollen- or stage-specific expression [22]. With the use of microarray, Chambers and Shuai detected 26 miRNAs which showed significant differences in expression between mature pollen and inflorescence in Arabidopsis. They confirmed the expression of 22 miRNAs in mature pollen by using real-time PCR, with most of miRNAs being expressed in low abundance [23]. Grant-Downton et al. detected 33 different microRNA families in mature pollen, and several showed pollen-enriched expression compared with leaves, such as miR156, miR2939, miR158, and miR845 [24]. Previous studies demonstrated the existence of miRNAs in pollen or inflorescence. However, studies must be conducted to investigate whether miRNAs participate in the pollen development and to determine their corresponding functions.
In this study, two small RNA libraries were constructed from the flower buds of the sterile line Bcajh97-01A (A line) and the fertile line Bcajh97-01B (B line) plants. A total of 24 known miRNAs were detected, 54 conserved miRNAs and 25 pairs of novel miRNA/miRNA* were identified. Meanwhile, 18 differentially expressed miRNAs were identified by comparing their expression abundances in the two libraries. Results from qRT-PCR agreed with those from high-throughput sequencing. To search for the target genes of identified miRNAs, degradome sequencing was conducted with the inflorescences of the B line plants. A total of 15 targets were predicted to be cleaved by seven miRNAs. Our study provides clues to explore miRNA group involved in pollen development and the interactions between miRNAs and their targets.
Results
Analysis of small RNA library data sets and the small RNA profile
To identify miRNAs involved in pollen development in the male sterile line ‘Bcajh97-01A’ and the male fertile line ‘Bcajh97-01B’, two independent small RNA libraries from the flower buds collected from the two lines were sequenced by using Illumina Solexa high-throughput sequencing technology. The two libraries generated a total of 6998586 and 6792888 raw reads, respectively (Table 1). The raw reads of the two libraries were uploaded to SRA database of NCBI and two accession numbers were obtained, which were SRX462325 and SRX464860. After removing the reads because 3ADT was not found, reads <15 bases, and junk reads, 5098547 and 5309119 sequences were obtained with lengths that range from 15 nt to 30 nt, respectively. After further filtering the RFam (rRNA, tRNA, snRNA, snoRNA, and other Rfam RNAs) and Repbase sequences, a total of 4540620 and 4667617 mappable small RNA sequences, respectively, were obtained (Table 1). The length distributions of small RNAs were very similar between the two libraries (Figure 1). In general, the majority of the small RNAs ranged from 21 nt to 24 nt in size. The 24 nt small RNAs in total sequence reads were the most dominant, followed by 21 nt small RNAs (Figure 1).
Identification of known miRNAs
To identify known miRNAs in Brassica campestris, all mappable small RNA sequences were compared with the known plant miRNAs in the miRBase database. A total of 24 small RNAs that have the same sequences with the known bra-miRNAs in miRBase were identified (i.e., 24 known Brassica campestris miRNAs were identified). The numbers of reads of the 24 known miRNAs in the two small RNA libraries from flower buds of A line and B line are listed in Additional file 1: Table S2 Among the 24 known miRNAs, bra-miR159a, bra-miR160a-5p, bra-miR171e, bra-miR1885b, and bra-miR5724 showed very high expression levels.
Novel miRNA on the other arm of known pre-miRNA
The advent of high-throughput sequencing technology has given rise to the discovery that a great number of miRNAs and miRNAs* are simultaneously present on two arms of pre-miRNA secondary structures. miRNA and miRNA* are renamed miRNA-3p or miRNA-5p, which indicates their locations in the 5′ arm or 3′ arm of pre-miRNA secondary structures. Through high-throughput sequencing, eight novel miRNAs on the other arm of known pre-miRNAs were identified. miRNA sequences and the corresponding number of reads in flower buds of the A line and B line are listed in Table 2.
Identification of new conserved miRNA families and new miRNA members
To identify conserved miRNAs in Brassica campestris, all mappable small RNAs were mapped to the Brassica campestris genome sequences and known plant miRNAs in miRBase. If the small RNAs can exactly map to Brassica campestris genome sequences and can also match known plant miRNAs with no more than three mismatches, these small RNAs were classified as candidate conserved miRNAs. Five criteria described in the materials and methods were mainly used to strictly screen the candidate conserved miRNAs. As a result, 54 miRNAs (27 pairs of miRNAs) that belong to 15 families were identified (Additional file 1: Table S3). These 54 miRNAs have not been previously reported as bra-miRNAs in miRBase and show high sequence similarity to some of the known plant miRNAs. Most of these miRNAs were 21 nt long, with only few miRNAs having lengths of 20, 22, or 23 nt; this characteristic is common in most plant species. For the bra-miR156 family, eight members were obtained by using deep sequencing. Two family members (bra-miR168a and bra-miR168b) of bra-miR168 were identified. Two pairs of miRNAs that belong to bra-miR168a, with one pair being bra-miR168a-1-p5 and bra-miR168a-1-p3, the other pair being bra-miR168a-2-p5 and bra-miR162a-2-p3, were identified. The two pairs of bra-miR168a shared the same mature sequences. However, they were from different precursors, i.e., they came from different loci of the Brassica campestris genome. The two precursor sequences of bra-miR168a were highly similar with each other. These two pairs of miRNAs were called sub-members. This type of sub-member was also observed in the bra-miR395 family. Four sub-members (bra-miR395a-1, bra-miR395a-2, bra-miR395a-3, bra-miR395a-4) were identified for bra-miR395a. This phenomenon suggests that some highly similar MIRNA gene might be produced by a replication event from one origin sequence to another one, which results in more copies of the miRNA group. Except the abovementioned three miRNA families, only one miRNA member was identified for the rest of miRNA family. At the same time, six new miRNA members that belonged to three known miRNA families were discovered in the two small RNA libraries (Additional file 1: Table S4).
Identification of novel miRNAs
To predict novel miRNAs in Brassica campestris, all the mappable small RNAs were blasted to the Brassica campestris genome sequence in Brassica database and plant known miRNAs in miRBase. The small RNAs that exactly map to the genome sequence but not the plant known miRNAs were classified as candidate novel miRNAs. To increase predictive accuracy, five criteria described in the materials and methods were mainly used to search for novel miRNAs. Novel miRNAs were discovered in pairs since the miRNA/miRNA* criterion was used. As a result, 25 pairs of novel miRNAs/miRNA* that belong to 23 miRNA families were identified in this study (Table 3). The bra-miRn1 had two sub-members (bra-miRn1-1 and bra-miRn1-2), as well as bra-miRn10 (bra-miRn10-1 and bra-miRn10-2). The lengths of mature miRNAs were distributed in the range of 21 nt to 24 nt. The MFE of these predicted pre-miRNAs ranged from -37 kcal/mol to -209.8 kcal/mol. The MFEI ranged from 0.9 to 2.1, with an average of 1.3, which is consistent with the characteristics of miRNA. Most of these novel miRNAs were expressed in the flower buds of the A line and B line, but the expression levels were very low. bra-miRn22-3p showed obviously high expression abundance in the flower buds of the A line and B line, which was considerably higher than expression levels of other miRNAs. The information about the numbers of reads and the sequence characteristics of all the identified miRNAs by using high-throughput sequencing are summarized in Additional file 2: Table S5. The hairpin structures for precursors of bra-miRn9 and bra-miRn10-1 are used as examples in Figure 2.
Expression profiling of differentially expressed miRNAs in the flower buds of A line and B line
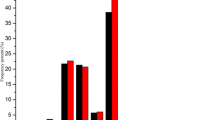
Throughout all the identified conserved and novel miRNAs, 18 differentially expressed miRNAs with more than two-fold relative change between the flower buds of A line and B line were identified in high-throughput sequencing (Figure 3). The relative expression level was calculated based on the normalized number of sequence reads of these miRNAs in small RNA libraries from flower buds of A line and B line. Among the 18 differentially expressed miRNAs, 15 miRNAs were up-regulated in flower buds of the B line. The remaining three miRNAs (bra-miR391a-p3, bra-miR390a-p5, and bra-miR168a-p5) showed higher expression levels in the flower buds of A line than in the B line (Figure 3). For all 15 miRNAs enriched in flower buds of the B line, bra-miR824-p3 showed the highest relative expression level (7.23-fold). For the three miRNAs enriched in flower buds of the A line, the differential expression of bra-miR168a-p5 was the most obvious (2.80-fold). qRT-PCR was conducted to verify the expression profile of the 18 differentially expressed miRNAs in deep sequencing. The results of qRT-PCR largely agreed with the deep sequencing results (Figure 3). In qRT-PCR, two more miRNAs (bra-miR391a-p3 and bra-miR168a-p5) were found to be up-regulated in the B line, which were enriched in the A line based on deep sequencing analysis. bra-miR390a-p5 was also up-regulated in the A line based on qRT-PCR analysis. In addition, expression profiles of these 18 miRNAs were presented in terms of the number of reads in flower buds of the A line and B line libraries (Figure 4). The number of normalized reads of bra-miR159a was very high, followed by bra-miR160a-5p, P-bra-miR319b-p3, and bra-miR168a-p5. P-bra-miR319b-p3 was predicted to be the 3′ arm miRNA of bra-miR319b. For all the identified miRNAs in this study, 5′ arm and 3′ arm miRNAs, namely, miRNA and miRNA*, were both detected, except for P-bra-miR319b-p3. Therefore, a “P” was added to bra-miR319b-p3, which denotes “predicted”. The two members of bra-miR168 family, namely, bra-miR168a-p3 and bra-miR168b-p3, were both up-regulated in flower buds of the B line. bra-miR824-p3 and bra-miR824 were consistently up-regulated in flower buds of the B line, which indicates that different members can have similar expression patterns.
Relative expression analysis of miRNAs in the flower buds of the A line and B line by high-throughput sequencing and qRT-PCR analysis. Relative expression level was normalized to the expression level of 5.8SrRNA in qRT-PCR. All qRT-PCR reactions were prepared in triplicate for each sample. Left indicates the miRNA relative expression level generated from the high-throughput sequencing. Right indicates the miRNA relative expression level obtained by using qRT-PCR analysis.
Expression profiles of differentially expressed miRNAs in flower buds of the A line and B line by high-throughput sequencing. The Y axis represents the number of reads of miRNAs detected in small RNA libraries from flower buds of A line (black bars) and B line (red bars) by using high-throughput sequencing.
Identification of miRNA target genes in Brassica campestris by using degradome analysis
Target identification of the miRNAs is important to further understand the potential regulatory role and biological function of a miRNA. In this study, degradome sequencing was used to search for the target genes of identified miRNAs in Brassica campestris. A total of 15 targets were predicted to be cleaved by seven miRNAs (Table 4). The seven miRNAs were bra-miR156, bra-miR159, bra-miR161, bra-miR172, bra-miR824, bra-miR1885, and bra-miRn4. bra-miR156 was identified in this study and was found to be highly conserved in plant species. Three targets that encode squamosa promoter binding protein (SBP) transcription factors were identified for bra-miR156. bra-miR159 and bra-miR172 were highly conserved miRNAs. They respectively targeted three and two transcription factors that belong to MYB domain protein family and AP2-like factor, respectively. bra-miR161 and bra-miR824 were both Cruciferae-specific miRNAs. bra-miR161 was identified in the present deep sequencing analysis and was found to target two pentatricopeptide repeat (PPR)-containing protein family genes. bra-miR824 is a known miRNA, which was predicted to cleave an AGAMOUS-like transcription factor. bra-miR1885 is a known miRNA that is only reported in Brassica campestris and might be specific only to Brassica. bra-miR1885 was predicted to target two genes encoding disease resistance protein (TIR-NBS-LRR class). bra-miRn4 is a novel miRNA that was predicted to cleave two genes that encode unknown proteins. Most of the identified targets are generally homologous to target genes found in Arabidopsis (Table 4). Unfortunately, for most miRNAs identified through deep sequencing, including conserved miRNAs and novel miRNAs, their target genes could not be detected in the present degradome analysis. In the above analysis, 15 target genes were identified for seven miRNA families. Among the 15 targets, nine were transcription factors. For each miRNA family, one target was chosen and a t-plot was constructed (Figure 5). In t-plots, the cleavage site for each miRNA:mRNA alignment is shown. The t-plots for the remaining eight targets are illustrated in Additional file 3: Figure S1.
Target plots (t-plots) of miRNAs targets confirmed by using degradome sequencing. A degradome cDNA library was constructed from the inflorescences of the B line and subjected to Illumina sequencing. A t-plot (top) and the corresponding miRNA:mRNA alignment (bottom) are shown for each of the seven target transcripts. In t-plots, the red arrows indicate the miRNA-directed cleaved transcript. The X axis indicates the nucleotide position in target cDNA. The Y axis indicates the number of reads of cleaved transcripts detected in the degradome cDNA library. The solid lines and dots in miRNA:mRNA alignments indicate matched and mismatched base pairs, respectively, and the red arrows and letters indicate the cleavage sites.
Discussion
Characteristics of conserved and non-conserved miRNAs in plants
In plants, many miRNAs seems to be universally expressed among diverse angiosperms, such as miR156, miR159, miR160, miR162, miR171, miR172 and so on. Among them, a small number of miRNAs have also been detected in bryophyte, lycopod, gymnosperm, such as miR156, miR319 [25]. However, there are a large number of miRNAs which are just present in a few species, even in only one species. For example, miR415, miR416, miR417, and miR418 have only been detected in Arabidopsis and Oryza sativa[26, 27]. miR1885 and miR5718 have just been identified in Brassica campestris[28, 29]. Considering that some miRNAs are widespread, while others distribute in limited plant species, miRNAs are classified into two categories, ‘conserved’ miRNAs and ‘non-conserved’ miRNAs. In previous reports, the term ‘conserved’ miRNAs are mainly used when the miRNAs are present throughout at least one major ancient clade of land plants, for example angiosperms. ‘Non-conserved’ miRNAs are defined as those with a limited phylogenetic distribution and characterized by primarily being single-copy genes [25]. In present years, with the development of high-throughput sequencing, a high proportion of non-conserved miRNAs have been identified in many species, such as larch [30], Populus euphratica[31], rice [32], maize [33], and soybean [34]. In our study, 15 conserved miRNA families, which had not been previously reported in Brassica campestris but reported in other plant species, were identified. Meanwhile, 25 pairs of novel miRNA/miRNA* were identified according to strict miRNA/miRNA* identification criteria. They are likely to be Brassica campestris-specific miRNAs, of course which are classified into non-conserved miRNAs.
Previous reports have indicated that non-conserved miRNAs are often species specific, weakly expressed, and encoded by single loci [35], while highly conserved miRNAs are widespread, highly expressed, and most of them have more than one family member [36].Our results were just in accord with the previous results. In our study, the numbers of reads of the 25 pairs of novel miRNA/miRNA* were extremely low, except for bra-miRn22-3p. Most of their read numbers were less than 100. However, the read numbers of the conserved miRNAs were very high. Most of their read numbers were more than 1000. bra-miR159 had the highest number of reads, which was close to 45000. A similar phenomenon was also observed in Arabidopsis lyrata[35, 37]. In addition, in our study, except for bra-miRn1 and bra-miRn10 families, only one member was identified for the rest of 21 novel miRNA families, while most of the conserved miRNA families have more than two family members. Our results indicates that compared with conserved miRNAs, most of the novel miRNAs were encoded by a single locus., which was consistent with the previous study [35]. In general, all of the previous studies and our results verify that conserved and non-conserved miRNAs present nearly opposite characters. As for why they show so different characters, more research on evolution and functional diversification of MIRNA genes will be helpful to elucidate it.
Diverse miRNAs are present in pollen and they are possibly involved in pollen development
Previous studies have demonstrated that many miRNAs exist in Arabidopsis mature pollen. In 2009, Grant-Downton et al. detected 33 known miRNAs in Arabidopsis pollen by using 454 sequencing technology [24]. In the same year, Chambers and Shuai verified that many miRNAs are expressed in the pollen and inflorescences of Arabidopsis using miRNA array [23]. Many studies have also been conducted in rice. In 2011, Wei et al. used deep sequencing technology to analyze the composition and expression patterns of miRNAs in developing pollen of rice, including uninucleate microspores, bicellular pollen and tricellular pollen, as well as sporophytic tissues. A total of 292 known miRNAs and 75 novel miRNAs were detected. Among the 292 known miRNAs, 103 were enriched in developing pollen and more than half of the novel miRNAs displayed pollen- or stage-specific expression. These pollen- or stage-specific miRNAs might function during pollen development process [22]. In our study, we identified 24 known miRNAs, 54 conserved miRNAs, and 25 pairs of novel miRNA/miRNA* in flower buds of the A line and B line plants. Among all these miRNAs, 18 differentially expressed miRNAs with more than two-fold relative change between flower buds of A line and B line were identified. Moreover, most of the 18 differentially expressed miRNAs were up-regulated in flower buds of B line. We speculate that these miRNAs might be involved in pollen development process. Previous studies and the present study demonstrate that diverse miRNAs exist in plant pollen and they might have potential regulatory roles in pollen development.
In 2009, Grant-Downton et al. demonstrated that several mainly sRNA pathway genes, including AGO family members, DCL1-4, HASTY, SERRATE, HEN1, and RDR family members, were expressed in unicellular microspores, bicellular pollen, tricellualr pollen, and mature pollen grains in Arabidopsis by RT-PCR analysis. At the same time, they succeeded in amplifying the pri-miRNAs, pre-miRNAs, and mature miRNAs from mature pollen cDNA, which verified that miRNA synthesis in pollen was going on as usual [38]. Their results further verify that miRNAs are not only present in pollen, but also may participate in the complex regulatory network of pollen development.
The application of degradome analysis have massively accelerated the research on the interactions of miRNAs and their target genes
Identification of target genes is the first and essential step to understand the regulatory roles of miRNAs. In plants, most miRNAs can perfectly or almost perfectly bind to their target mRNAs. Thus, bioinformatics prediction is a common approach for predicting miRNA target genes. In recent years, many computational tools have been developed for predicting plant miRNA targets, such as psRNATarget [39], Target-align [40], TAPIR [41], and PMRD [42]. 5′-modified RACE is frequently used to demonstrate miRNA targets. This method is easy to operate, and the results are reliable. However, 5′-modified RACE is based on the premise that the candidate target is already predicted and its mRNA sequence is known for primer design. 5′-modified RACE is not efficient for identifying a large number of candidate target genes. Fortunately, in recent years, degradome sequencing analysis [43], a new method for massively identifying target genes, is increasingly being developed for such applications. The application of this new method greatly accelerates the detection of miRNA targets.
In recent years, degradome sequencing has been used for large-scale target identification in many species, such as Populus euphratica[31], soybean [34], cucumber [44], and Brassica juncea[45]. In the present study, degradome sequencing was performed and finally a large number of candidate target genes were detected. In order to confirm the reliable candidate targets, two criteria described in the materials and methods were mainly used. Finally, a total of 15 targets were chosen and predicted to be cleaved by seven miRNA families. Nine of the 15 targets were transcription factors. They were squamosa promoter binding (SBP) transcription factors (Bra032822, Bra010949, Bra030041), MYB transcription factors (Bra034842, Bra002042, Bra035547), AP2-like transcription factors (Bra017809, Bra011741), AGAMOUS-like transcription factors (Bra011509), which were targeted by miR156, miR159, miR172, miR824 families, respectively. These target genes have been reported playing an important role in plant growth and development. miR172, which targets AP2-like transcription factors, has been implicated in the regulation of flowering time and floral organ identity in maize and Arabidopsis[16, 17]. In 2009, Wu et al. indicated that miR156 and miR172 regulated the development transition from juvenile to adult [46]. In the next year, Xing et al. concluded that fully fertile Arabidopsis flowers required the action of multiple miR156/7-targeted SPL genes in concert with SPL8, otherwise semi-sterile or fully sterile would emerge [47]. The results suggested that miR156/7 and their targets, namely SBP transcription factors, might participate in gametophyte development. miR159 and its target genes, namely MYB-like genes, were proved to inhibit growth and promote programmed cell death in Arabidopsis[48]. In addition to targeting transcription factors, the remaining 6 target genes were also shown to be involved in other biological processes. For example, miR1885 and its two targets were involved in disease resistance [29]. miR161 and bra-miRn4 both had two candidate target genes and their functions in plants were unknown yet. In summary, degradome analysis has greatly accelerated the identification of miRNA targets. Meanwhile, it speeds up the research on miRNA/target interactions.
Conclusion
In this study, a large number of miRNAs were identified in Brassica campestris ssp. chinensis, including 8 novel miRNAs on the other arm of known pre-miRNAs, 54 conserved miRNA families, 8 new miRNA members that belong to 8 known miRNA families, and 25 pairs of novel miRNA/miRNA*. Meanwhile, by analyzing the sequencing reads of miRNAs, we found that there were18 miRNAs differentially expressed between the flower buds of A line and B line, and 15 of them were up-regulated in flower buds of B line. This result was validated by using qRT-PCR analysis. By using degradome sequencing, a total of 15 targets were identified for 7 miRNA families, which reveal interaction between miRNAs and targets. In a word, the identification of plenty of miRNAs has greatly enriched the existing miRNA group in Brassica campestris. More importantly, the present study might provide valuable clues for exploring miRNA-mediated regulatory networks during pollen development.
Methods
Plant materials, sample collection, and total RNA extraction
A genic male sterile system in Chinese cabbage-pak-choi (Brassica campestris ssp. chinensis, syn. Brassica rapa ssp. chinensis), named ‘Bcajh97-01A/B’ was used in this study. ‘Bcajh97-01A’ is a male sterile line that lacks mature pollen, and its male sterility is controlled by a pair of nuclear recessive genes. ‘Bcajh97-01B’ is the fertile line that generates normal mature pollen and is the maintainer line of ‘Bcajh97-01A’. The ‘Bcajh97-01A/B’ sister line system has been developed through continuous backcrossing within the population for several generations [49]. The progenies of ‘Bcajh97-01A/B’ line regularly segregate into sterile and fertile types during reproduction at a ratio of 1:1. The characteristics of male sterility can be steadily maintained.
All plant materials were grown in the experimental farm of Zhejiang University. In this study, three kinds of samples were harvested at the flowering stage from ‘Bajh97-01A/B’ plants, which were the mixture of flower buds from ‘Bcajh97-01A’ and ‘Bcajh97-01B’ and the inflorescences from ‘Bcajh97-01B’ , respectively. Each kind of sample was collected from 10 different plants and subsequently mixed, flash frozen in liquid nitrogen, and stored at -80°C until total RNA isolation. Total RNAs of the three kinds of samples were extracted by using mirVana kit (Ambion, USA) according to the manufacturer’s instructions.
Small RNA library construction and sequencing
Small RNA library construction was conducted by using Illumina TruSeq Small RNA Preparation Kit following the manufacturer’s instructions (LC Sciences, Hangzhou, China). The general process is as follows: first, the total RNA was ligated to RNA 3′ and RNA 5′ adapters. Second, reverse transcription followed by PCR was performed to create cDNA constructs based on the small RNAs ligated with 3′ and 5′ adapters. Third, small cDNA fractions that range from 22 nt to 30 nt in length were isolated by using 6% denaturing polyacrylamide gel electrophoresis. Fourth, cDNA construct was purified, and the library was validated.
The purified cDNA library was used for cluster generation on Illumina’s Cluster Station and subsequently sequenced on Illumina GAIIx (Illumina, Inc., Santa Clara, CA) following the manufacturer’s instruction on running the instrument. Raw sequencing reads were obtained by using related Illumina’s analysis software. The ACGT101-miR program (version 4.2; LC Sciences) was used for standard sequencing data analysis.
Identification of conserved and novel miRNAs
After the raw sequence reads were extracted, adapter sequences, impurities, and sequences beyond 15 nt to 30 nt were filtered. The remaining sequences that range from 15 nt to 30 nt in length were used for miRNA prediction by using the ACGT101-miR program (version 4.2; LC Sciences). First, the sequences were blasted to the RFam database (RFam: rRNA, tRNA, snRNA, snoRNA, and other non-coding RNAs), repeat sequences, and mRNAs. Matched sequences were discarded. The sequences were then compared with the Brassica campestris genome sequences downloaded from the Brassica database (http://brassicadb.org/brad/). The unmatched sequences were filtered. Finally, the remaining sequences were mapped to all known plant miRNAs sequences to identify the conserved miRNAs in Brassica campestris from the miRBase database (version 19.0, http://www.mirbase.org/). Matched sequences with no more than three mismatches were considered as candidate conserved miRNAs. At the same time, the unmatched sequences were reserved as candidate novel miRNAs. To identify conserved or novel miRNAs in Brassica campestris, novel and conserved candidate miRNAs sequences were blasted against Brassica campestris genome sequences, and their flanking sequences in the genome were used to predict their secondary structures by using the mfold Web server (http://mfold.rna.albany.edu/?q=mfold/download-mfold) [50]. A potential miRNA precursor must be a non-coding sequence and must meet certain criteria. The first and the most important criterion is the miRNA/miRNA* criterion. Both a candidate miRNA and its corresponding reverse sequence, namely, the candidate miRNA* sequence, must be detected in the present high-throughput sequencing. Second, the candidate miRNA and miRNA* sequences must be found on the stem, and the number of mismatched bases between them must be less than four (four continuous mismatches are also not allowed). Third, within the miRNA/miRNA* duplex, the number of asymmetric bulges must be one or fewer, and the number of bases in the asymmetric bulges must fewer than two. Fourth, the miRNA and miRNA* should be located in opposite stem-arms and form a duplex with two nucleotide 3′ overhangs [51]. Fifth, the potential miRNA precursor must have higher negative minimal folding energy (MFE) and minimal folding energy indexes (MFEI), with the MFEI > 0.8, to distinguish from other small RNAs [52]. After the above strict screening, conserved and novel miRNAs are identified in Brassica campestris.
Degradome library construction, data analysis, and target identification
A degradome library was constructed from the inflorescences of the fertile line (Bcajh97-01B) based on the method described by German et al. [43] and Addo-Quaye [53]. Briefly, poly(A)-enriched RNA was ligated to a 5′-RNA adapter with 3′ a EcoP15 I recognition site. Reverse transcription was performed to generate first-strand cDNA, followed by PCR amplification and EcoP15 I digestion. After digestion with EcoP15 I, a PAGE-gel was used to purify the EcoP15 I-cleaved fragments. The gel-purified products were ligated to a 3′-double-strand DNA adapter, followed by PAGE-gel purification to obtain the ligated products. PCR amplification was performed, and PAGE-gel was used for the third time to purify the corresponding gel bands containing the final products. Finally, the purified cDNA library was ready for deep sequencing (LC Sciences, Hangzhou, China).
The purified cDNA library was first used for cluster generation on Illumina’s Cluster Station and then sequenced on Illumina GAIIx. Raw sequencing reads were obtained by using Illumina’s Pipeline v1.5 software following sequencing image analysis by Pipeline Firecrest Module and base-calling by using Pipeline Bustard Module (LC Sciences, Hangzhou, China). A public software package, CleaveLand 3.0, was used for analyzing sequencing data [53, 54].
By degradome sequencing, a great many genes may be predicted as potential target genes. In this study, two criteria were mainly used to choose reliable genes as candidate targets. First, the cleavage site must be the 9th or 10th nucleotide of the target mRNA in the miRNA/target binding region. Second, the candidate targets must be homologous to corresponding A. thaliana targets [55]. The candidate target genes that meet the above criteria would be identified as targets.
Quantitative real-time PCR
Total RNA was extracted from the flower buds of the sterile line and the fertile line by using the mirVana kit (Ambion, USA). According to the procedures provided by a miRNA cDNA synthesis kit (TaKaRa, Japan), 1 μg of total RNA was polyadenylated with ATP by poly(A) polymerase. The poly(A)-tailed total RNA was reverse-transcribed by PrimeScript® RTase by using a universal adapter primer (containing oligo-dT). qRT-PCR analysis was carried out by using SYBR® Premix Ex TaqTM II (Perfect Real Time) (TaKaRa, Japan) on a Bio-Rad CFX96 machine. All reactions were performed in triplicate for each sample, and 5.8S rRNA was used as the internal control gene. Relative expression levels of miRNAs were quantified by using the 2-ΔΔCt method [56]. The primers used for qRT-PCR are listed in Additional file 1: Table S1.
References
McCormick S: Male gametophyte development. Plant Cell. 1993, 5: 1265-1275.
Goldberg RB, Beals TP, Sanders PM: Anther development: basic principles and practical applications. Plant Cell. 1993, 5: 1217-1229.
McCormick S: Control of male gametophyte development. Plant Cell. 2004, 16 (Suppl): S142-S153.
Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X: Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA. 2010, 107: 6100-6105. 10.1073/pnas.0912333107.
Honys D, Twell D: Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 2003, 132: 640-652. 10.1104/pp.103.020925.
Dobritsa AA, Geanconteri A, Shrestha J, Carlson A, Kooyers N, Coerper D, Urbanczyk-Wochniak E, Bench BJ, Sumner LW, Swanson R, Preuss D: A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 2011, 157: 947-970. 10.1104/pp.111.179523.
Huang L, Cao JS, Zhang AH, Ye YQ, Zhang YC, Liu TT: The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. J Exp Bot. 2009, 60: 301-313.
Liu L-C, Cao J-S, Yu X-L, Xiang X, Fei Y-J: Expression of an Antisense BcMF3 affects microsporogenesis and pollen tube growth in arabidopsis. Agri Sci China. 2006, 5: 339-345. 10.1016/S1671-2927(06)60059-X.
Liu LC, Xiang X, Cao JS: [BcMF4 gene, encoding a leucine-rich repeat protein, plays a role in male fertility in Chinese cabbage-pak-choi]. Yi Chuan. 2006, 28: 1428-1434.
Jiang J, Yu X, Miao Y, Huang L, Yao L, Cao J: Sequence characterization and expression pattern of BcMF21, a novel gene related to pollen development in Brassica campestris ssp. chinensis. Mol Biol Rep. 2012, 39: 7319-7326. 10.1007/s11033-012-1563-6.
Zhang Q, Huang L, Liu T, Yu X, Cao J: Functional analysis of a pollen-expressed polygalacturonase gene BcMF6 in Chinese cabbage (Brassica campestris L. ssp. chinensis Makino). Plant Cell Rep. 2008, 27: 1207-1215. 10.1007/s00299-008-0541-x.
Huang L, Ye Y, Zhang Y, Zhang A, Liu T, Cao J: BcMF9, a novel polygalacturonase gene, is required for both Brassica campestris intine and exine formation. Ann Bot. 2009, 104: 1339-1351. 10.1093/aob/mcp244.
Guo HS, Xie Q, Fei JF, Chua NH: MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005, 17: 1376-1386. 10.1105/tpc.105.030841.
Floyd SK, Bowman JL: Gene regulation: ancient microRNA target sequences in plants. Nature. 2004, 428: 485-486. 10.1038/428485a.
Wang JW, Park MY, Wang LJ, Koo Y, Chen XY, Weigel D, Poethig RS: miRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7: e1002012-10.1371/journal.pgen.1002012.
Aukerman MJ, Sakai H: Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003, 15: 2730-2741. 10.1105/tpc.016238.
Chen X: A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004, 303: 2022-2025. 10.1126/science.1088060.
Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M: Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem. 2007, 282: 16369-16378. 10.1074/jbc.M700138200.
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP: Prediction of plant microRNA targets. Cell. 2002, 110: 513-520. 10.1016/S0092-8674(02)00863-2.
Sunkar R, Chinnusamy V, Zhu J, Zhu JK: Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12: 301-309. 10.1016/j.tplants.2007.05.001.
Katiyar-Agarwal S, Jin H: Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010, 48: 225-246. 10.1146/annurev-phyto-073009-114457.
Wei LQ, Yan LF, Wang T: Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 2011, 12: R53-10.1186/gb-2011-12-6-r53.
Chambers C, Shuai B: Profiling microRNA expression in Arabidopsis pollen using microRNA array and real-time PCR. BMC Plant Biol. 2009, 9: 87-10.1186/1471-2229-9-87.
Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H: MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics. 2009, 10: 643-10.1186/1471-2164-10-643.
Axtell MJ, Bowman JL: Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008, 13: 343-349. 10.1016/j.tplants.2008.03.009.
Wang XJ, Reyes JL, Chua NH, Gaasterland T: Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004, 5: R65-10.1186/gb-2004-5-9-r65.
Jones-Rhoades MW, Bartel DP, Bartel B: MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006, 57: 19-53. 10.1146/annurev.arplant.57.032905.105218.
Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, Prins M, van Tunen A, He Y: Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot. 2012, 63: 1025-1038. 10.1093/jxb/err337.
He XF, Fang YY, Feng L, Guo HS: Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008, 582: 2445-2452. 10.1016/j.febslet.2008.06.011.
Zhang J, Zhang S, Han S, Wu T, Li X, Li W, Qi L: Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta. 2012, 236: 647-657. 10.1007/s00425-012-1643-9.
Li B, Qin Y, Duan H, Yin W, Xia X: Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot. 2011, 62: 3765-3779. 10.1093/jxb/err051.
Heisel SE, Zhang Y, Allen E, Guo L, Reynolds TL, Yang X, Kovalic D, Roberts JK: Characterization of unique small RNA populations from rice grain. PLoS One. 2008, 3: e2871-10.1371/journal.pone.0002871.
Zhao M, Tai H, Sun S, Zhang F, Xu Y, Li WX: Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One. 2012, 7: e29669-10.1371/journal.pone.0029669.
Zeng QY, Yang CY, Ma QB, Li XP, Dong WW, Nian H: Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 2012, 12: 182-10.1186/1471-2229-12-182.
Ma ZR, Coruh C, Axtell MJ: Arabidopsis lyrata Small RNAs: Transient MIRNA and Small Interfering RNA Loci within the Arabidopsis Genus. Plant Cell. 2010, 22: 1090-1103. 10.1105/tpc.110.073882.
Cuperus JT, Fahlgren N, Carrington JC: Evolution and functional diversification of MIRNA genes. Plant Cell. 2011, 23: 431-442. 10.1105/tpc.110.082784.
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP: A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20: 3407-3425. 10.1101/gad.1476406.
Grant-Downton R, Hafidh S, Twell D, Dickinson HG: Small RNA pathways are present and functional in the angiosperm male gametophyte. Mol Plant. 2009, 2: 500-512. 10.1093/mp/ssp003.
Dai X, Zhao PX: psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011, 39: W155-W159. 10.1093/nar/gkr319.
Xie F, Zhang B: Target-align: a tool for plant microRNA target identification. Bioinformatics. 2010, 26: 3002-3003. 10.1093/bioinformatics/btq568.
Bonnet E, He Y, Billiau K, Van de Peer Y: TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010, 26: 1566-1568. 10.1093/bioinformatics/btq233.
Zhang Z, Yu J, Li D, Liu F, Zhou X, Wang T, Ling Y, Su Z: PMRD: plant microRNA database. Nucleic Acids Res. 2010, 38: D806-D813. 10.1093/nar/gkp818.
German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ: Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008, 26: 941-946. 10.1038/nbt1417.
Mao W, Li Z, Xia X, Li Y, Yu J: A combined approach of high-throughput sequencing and degradome analysis reveals tissue specific expression of microRNAs and their targets in cucumber. PLoS One. 2012, 7: e33040-10.1371/journal.pone.0033040.
Yang J, Liu X, Xu B, Zhao N, Yang X, Zhang M: Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genomics. 2013, 14: 9-10.1186/1471-2164-14-9.
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS: The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009, 138: 750-759. 10.1016/j.cell.2009.06.031.
Xing S, Salinas M, Hohmann S, Berndtgen R, Huijser P: miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell. 2010, 22: 3935-3950. 10.1105/tpc.110.079343.
Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA: The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154: 757-771. 10.1104/pp.110.160630.
Huang L, Cao J, Ye W, Liu T, Jiang L, Ye Y: Transcriptional differences between the male-sterile mutant bcms and wild-type Brassica campestris ssp chinensis reveal genes related to pollen development. Plant Biology. 2008, 10: 342-355. 10.1111/j.1438-8677.2008.00039.x.
Zuker M: Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31: 3406-3415. 10.1093/nar/gkg595.
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK: Criteria for annotation of plant MicroRNAs. Plant Cell. 2008, 20: 3186-3190. 10.1105/tpc.108.064311.
Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA: Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci. 2006, 63: 246-254. 10.1007/s00018-005-5467-7.
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ: Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008, 18: 758-762. 10.1016/j.cub.2008.04.042.
Addo-Quaye C, Miller W, Axtell MJ: CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009, 25: 130-131. 10.1093/bioinformatics/btn604.
Jiang JX, Jiang JJ, Yang YF, Cao JS: Identification of microRNAs potentially involved in male sterility of Brassica campestris ssp chinensis using microRNA array and quantitative RT-PCR assays. Cell Mole Biol Lett. 2013, 18: 416-432. 10.2478/s11658-013-0097-9.
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001, 25: 402-408. 10.1006/meth.2001.1262.
Acknowledgements
This work was supported by the National Program on Key Basic Research Projects (No. 2012CB113900), the Natural Science Foundation of China (No. 31071805) and the Key Sci-Technology Project of Zhejiang Province (No. 2010C12004) and the Breeding project of the Science-Technology Foundation of Zhejiang provice (Grant No. 2012C12903).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JXJ and JSC designed the study. JXJ performed the experiments, analyzed the data, and drafted the manuscript. YL and MLL assisted with bioinformatic analysis and aided in writing the manuscript. ZMM aided in performing the experiments. All authors carefully checked and approved this version of the manuscript.
Electronic supplementary material
12864_2013_5775_MOESM1_ESM.pdf
Additional file 1: Table S1: Primer sequences used for quantitative RT-PCR analyses. Table S2. Identification of known miRNAs. Table S3. Identification of new conserved miRNAs in Brassica campestris. Table S4. Identification of new miRNA family members by high-throughput sequencing. (PDF 34 KB)
12864_2013_5775_MOESM2_ESM.xls
Additional file 2: Table S5: Information about the numbers of reads and the sequence characteristics of all the identified miRNAs by using high-throughput sequencing. (XLS 87 KB)
12864_2013_5775_MOESM3_ESM.pdf
Additional file 3: Figure S1: Target plots (t-plots) of miRNAs targets confirmed by using degradome sequencing in Brassica campestris.(PDF 60 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Jiang, J., Lv, M., Liang, Y. et al. Identification of novel and conserved miRNAs involved in pollen development in Brassica campestris ssp. chinensi s by high-throughput sequencing and degradome analysis. BMC Genomics 15, 146 (2014). https://doi.org/10.1186/1471-2164-15-146
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-15-146