Abstract
Background
Interferon inducible transmembrane proteins (IFITMs) have diverse roles, including the control of cell proliferation, promotion of homotypic cell adhesion, protection against viral infection, promotion of bone matrix maturation and mineralisation, and mediating germ cell development. Most IFITMs have been well characterised in human and mouse but little published data exists for other animals. This study characterised IFITMs in two distantly related marsupial species, the Australian tammar wallaby and the South American grey short-tailed opossum, and analysed the phylogeny of the IFITM family in vertebrates.
Results
Five IFITM paralogues were identified in both the tammar and opossum. As in eutherians, most marsupial IFITM genes exist within a cluster, contain two exons and encode proteins with two transmembrane domains. Only two IFITM genes, IFITM5 and IFITM10, have orthologues in both marsupials and eutherians. IFITM5 arose in bony fish and IFITM10 in tetrapods. The bone-specific expression of IFITM5 appears to be restricted to therian mammals, suggesting that its specialised role in bone production is a recent adaptation specific to mammals. IFITM10 is the most highly conserved IFITM, sharing at least 85% amino acid identity between birds, reptiles and mammals and suggesting an important role for this presently uncharacterised protein.
Conclusions
Like eutherians, marsupials also have multiple IFITM genes that exist in a gene cluster. The differing expression patterns for many of the paralogues, together with poor sequence conservation between species, suggests that IFITM genes have acquired many different roles during vertebrate evolution.
Similar content being viewed by others
Background
The human nter eron nducible rans embrane (IFITM) genes were originally identified by their differential response to stimulation by interferon [1, 2], mediated by interferon-stimulated response elements (ISREs). The family consists of five genes (IFITM1 (= Leu-13 or 9 27), IFITM2 (1-8D) IFITM3 (1-8U) IFITM5 and IFITM10) all on chromosome 11 (Figure 1). All five genes encode proteins with 125–133 amino acids and two transmembrane domains and IFITM1, -2 and −3 have ISREs just 5' to their start codons [3]. Numerous biological roles have been attributed to IFITM1, -2 and −3 but little is known of the roles of IFITM5 and IFITM10 in humans. IFITM1 is expressed by leukocytes and endothelial cells, has anti-proliferative effects and promotes homotypic cell adhesion [4–6]. IFITM3 also inhibits cell proliferation [7], while IFITM2 induces both cell cycle arrest and subsequent p53-independent apoptosis in numerous cell lines [8]. IFITM proteins are also important components of the interferon-mediated innate immune system, and interest in these proteins has increased recently with the discovery that IFITM1, -2 and −3 offer protection against numerous viruses, including Influenza A, Dengue and HIV [9–11]. They may also act as tumor suppressors because of their ability to control the cell cycle. Supporting this idea, cells transitioning from a normal to a pre-malignant state often exhibit abnormal IFITM expression [12].
Arrangement of IFITM gene clusters in the tammar, opossum, human and mouse. Arrows indicate the direction of transcription. The paler bar for IFITMA in the opossum indicates that there is not enough information available to determine whether the contig containing IFITMA is continuous with the cluster containing IFITMB1, -B2 and −5.
The Ifitm gene family has been well characterised in mice. Ifitm3 (also known as Mil1 and Fragilis), the first murine Ifitm gene to be characterised, was identified by a screen for genes expressed specifically in early primordial germ cells (PGCs) [13] and also in a subtractive cDNA screen comparing migrating PGCs with inner cell mass cells [14]. Ifitm3 is on mouse chromosome 7, together with Ifitm1 (Mil2, Fragilis 2), Ifitm2 (Mil3, Fragilis 3), Ifitm5 (Fragilis 4), Ifitm6 (Fragilis 5) [15] and Ifitm10 (also known as 6330512M04Rik), whereas Ifitm7 (Mil4) is on mouse chromosome 16 [14]. Murine Ifitm genes encode proteins with 104–144 amino acids and two transmembrane domains. Ifitm1 -3 and −6 also have ISREs [14, 15]. Mouse Ifitm1 -2 and −3 genes are not respectively orthologous to human IFITM1 -2 and −3 genes, and sequence comparisons between human and mouse Ifitm genes suggest that duplication of the Ifitm genes occurred independently in human and mouse lineages [15].
Ifitm5 was renamed bone restricted Ifitm-like protein (Bril) because its expression in both humans and mice is restricted to bone, especially osteoblasts, and functional studies in mice suggest a role in bone matrix mineralisation and maturation [16]. Nothing is known of the functions of Ifitm6 -7 or −10. During murine embryonic development, Ifitm1 is expressed in the extraembryonic and embryonic mesoderm until the mesoderm differentiates [14, 15]. Ifitm2 is ubiquitously expressed from E8.5, and Ifitm3 expression mirrors the development of the germ cell lineage: it is expressed in the proximal epiblast from E5.5 and its expression is gradually restricted to PGCs as gastrulation proceeds [13–15]. Ifitm5 and −6 are not expressed in the embryo between E5.5 and E9.5 [15] but Ifitm5 is expressed from E14.5 in developing bone [16].
Ifitm3 expression delineates cells competent to become PGCs as early as E6.25 and it may have a role in germ cell development, possibly by promoting the formation of a discrete cell population that distinguishes presumptive PGCs from somatic cells via homotypic cell adhesion [13]. Ifitm1 and −3 may also control PGC migration. Application of ectopic Ifitm1 and −3 or silencing of Ifitm1 via short hairpin RNA (shRNA) knockdown in embryos results in abnormal PGC migration [17]. Ifitm1 could also be required for somite epithelialisation and formation of paraxial mesoderm, as these processes are defective when Ifitm1 is silenced in vivo by RNA interference [18]. Knockdown of Ifitm5 in osteoblast cell lines in vitro results in reduced bone mineralisation, implicating Ifitm5 in matrix mineralisation and maturation [16]. Mice homozygous null for Ifitm5 have smaller skeletons than heterozygous or wild type mice but their relatively mild phenotype suggests that the function of Ifitm5 can be compensated for by some other factor [19]. The possibility of redundancy of Ifitm genes is supported by another study in which the locus containing the Ifitm cluster (Ifitm1 -2 -3 -5 and −6) was deleted by floxing out a 120-kb region. The resulting mice, even those homozygous for the deletion, apparently developed normally and were fully fertile, suggesting that these Ifitm genes are not essential for normal germline development or any other developmental process [20] and that their roles can be compensated for by other genes.
The studies described above focused on human or mouse IFITMs, and aside from a rudimentary description of IFITM genes in the cow and rat [15], the only other published descriptions of IFITM genes are for the electric eel [21] and trout [22]. Interest in IFITMs in humans has increased with the discovery of their roles as anti-viral agents and as markers of cancer and inflammatory diseases [12]. Such roles have not been described in the mouse, in which the main focus on Ifitms has been with respect to their role(s) in the development of the germ cell lineage. The lack of sequence conservation of IFITMs between human and mouse suggests that their roles may vary between species. Marsupials diverged from eutherians approximately 160 million years ago [23] and the Australian and South American marsupials have evolved independently since the break-up of Gondwana approximately 80 million years ago [24]. Comparing marsupial and eutherian IFITM genes will provide new information on the conservation and evolution of this gene family in mammals. This study therefore describes IFITM genes in two marsupial species, the Australian tammar wallaby Macropus eugenii (a macropodid marsupial) and the South American short-tailed grey opossum Monodelphis domestica (a didelphid marsupial) and compares them with existing genomic databases.
Results
Marsupial IFITMs
The tammar and opossum genomes each have five IFITM paralogues (Figure 1). Tammar IFITM genes identified were IFITMA1 (Genbank accession number JQ254908), IFITMA2 (JQ254909), IFITMB (JQ254910), IFITM5 (JQ254911) and IFITM10 (JQ254912), encoding predicted protein sizes of 12.1 to 16.8 kDa. Opossum IFITMs identified were IFITMA, IFITMB1, IFITMB2 (Additional file 1: Table S 1), IFITM5 (XM_001363778, previously identified as interferon-induced transmembrane protein 5-like) and IFITM10 (XM_001367690, previously identified by Genbank as CD225 family protein FLJ76511-like), encoding predicted protein sizes ranging from 12.3 to 14.5 kDa. The only evidence of an opossum orthologue to tammar IFITMA is a short (796 nt) sequence (ti:515911565) in the opossum trace archives that contains both coding exons. There are several gaps in the current opossum genome assembly (Broad/monDom5) within the IFITM locus, so IFITMA may well be located within the IFITM cluster.
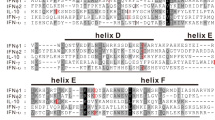
Marsupial IFITMs contain two exons and encode two transmembrane domains. The highest level of sequence conservation occurs within the first transmembrane domain and in between the two transmembrane domains (Figure 2A), similar to eutherian IFITM genes. Conservation of IFITM protein sequences both between and within the two marsupial species is quite low (22-38% similarity) with the exception of the comparisons listed in Table 1. The motif AGGAAATAGAAACT is an interferon stimulated response element (ISRE) in human IFITM1[2] and tammar IFITMA1 and IFITMA2 both have an identical putative ISRE (AGGAAATAGAAAG T) located close (299 and 193 nucleotides respectively) to the start of their open reading frames. No such motifs were identified in the other tammar IFITMs or in any of the opossum IFITMs, although minimal 5′ sequence is available for opossum IFITMA.
IFITM protein alignments. A, alignment of marsupial IFITM paralogues. B, C, alignments of IFITM10 and IFITM5 orthologues, respectively. Black shading represents identical amino acids conserved between all species, grey amino acids conserved in at least 75% of the species. Hatched bars underneath each alignment indicate the location of transmembrane domains.
Screening of the tammar BAC library with probes against IFITMA2 and IFITM10 yielded two positive clones. Sequencing of the IFITMA2-positive BAC clone showed that IFITMA1, -A2, -B and −5 are clustered together within a 74.5 kb region and share the same orientation (Figure 1). Using fluorescence in situ hybridisation (FISH), the IFITM cluster was localised to chromosome 5q and IFITM10 to chromosome 2p (Figure 3). The chromosomal location of the opossum IFITM genes is unknown but at least IFITMB1, -B2 and −5 are clustered, whereas IFITM10 is not within the main IFITM gene cluster. The location of IFITMA relative to the IFITM cluster is unknown.
Many of the human and some of the mouse IFITM genes have multiple transcripts, which differ either in their 5′, 3′ or both 5′ and 3′ untranslated regions (UTRs). Northern blots were attempted for each tammar IFITM gene but were unsuccessful (data not shown); probes to IFITMA1 and -A2 cross-reacted with each other whereas no distinct bands were obtained for IFITMB -5 or −10, despite the use of multiple tissue samples and different probes. Therefore each tammar IFITM gene was used to search the tammar transcriptome and EST (expressed sequence tag) databases [25]. No transcripts representing IFITM5 or IFITM10 were identified. Two sequences containing at least the second exon and the polyA signal of IFITMA2 and one for IFITMB were obtained and all supported the 3′ UTR identified by the BAC sequencing. There were four matches for IFITMA1, with two supporting a short 3′ UTR and the other two suggesting longer 3′ UTRs, only one of which contained a polyA signal. Thus, IFITMA1 appears to have several transcripts. A similar analysis for IFITMA2 also suggests several transcripts – whether these differ in the length of their 5′ or 3′ UTRs or contain additional exons upstream of the ones presented here is not known. The human and mouse genomes also each contain several IFITM processed pseudogenes. Searching the tammar genomic database [25] using tammar IFITM genes yielded one match for each gene, and in each instance this sequence contained an intervening intron.
The expression of IFITM genes in the tammar was examined in adult tissues and also in peri-gastrulation and fetal stages by RT-PCR. In adult organs, IFITMA1, -A2 and -B were widely expressed (Figure 4), whereas IFITM5 is only expressed in bone. IFITMA1, -A2 and -B were also the most widely expressed (temporally) IFITM genes during embryonic and fetal development (Figure 5), although IFITMB expression was very low in bilaminar (avascular) yolk sac (BYS). IFITM10 expression was also absent from the BYS and was not detectable until slightly later in development, during early somitogenesis. IFITM5 was not expressed at any pre-natal stage examined.
IFITMs in other taxa
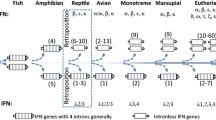
IFITM genes were identified in representatives from all seven major vertebrate classes (Figure 6, Table 2). There is no genome database available for urodelian amphibians and only a rudimentary one for chondrichthyes (cartilaginous fishes) so results in Figure 6 and Additional file 1: Table S 1 are based on EST data. An IFITM protein has also been described in the cartilaginous electric ray Torpdeo marmorata[21]. Only one small IFITM protein of approximately 30 amino acids was found for the lamprey (agnathan) and this aligns to IFITM1 and −3-like IFITM genes from the turkey, zebrafish and frog with a higher score than it does to any IFITM5 or −10 orthologue (data not shown). IFITM5 appears to have arisen in bony fish, and IFITM10 in tetrapods. The apparent lack of either of these genes in urodeles probably reflects the paucity of genomic data currently available.
Model for the evolution of IFITM genes in chordates. The IFITM gene (green line) in the lamprey is only a partial transcript. In some clades such as anurans, bony fish, marsupials and eutherians, there are several IFITM paralogues, presumably arising through gene duplication. Bony fish are the first class in which IFITM5 (blue line) is present, whereas IFITM10 (red line) is tetrapod-specific. Note that in the cartilaginous fishes and urodeles (for which only EST data is available), IFITM orthologues listed as not present (dotted lines) may in fact exist but have not been detected due to the lack of a comprehensive genomic database. Extensive searching of the platypus (a monotreme) genome yielded a full transcript for IFITM5, but only partial transcripts for IFITM and IFITM10. *this information is based only on EST data because a genomic database is not available.
A Neighbour joining phylogenetic tree constructed using IFITM amino acid sequences showed four distinct IFITM clusters (Figure 7). Two of these contained orthologues of IFITM5 and IFITM10 respectively from many different species, the third and fourth clusters contained the remaining eutherian and marsupial IFITMs respectively. The IFITM10 orthologues are the most highly conserved, exhibiting > 61% amino acid similarity between all species (increasing to > 85% similarity if frog IFITM10 is excluded). The IFITM5 orthologues share > 32% amino acid similarity (> 54% if fish and frog IFITM5s are excluded).
Phylogenetic tree for vertebrate IFITM proteins. The tree was constructed using the Neighbourhood joining method with 1000 bootstraps. Green shading denotes the cluster of general eutherian IFITMs, orange the marsupial IFITM cluster, pink the IFITM10 cluster and blue the IFITM5 cluster. Only IFITMs for which complete (or almost complete) protein sequence was available were included in this analysis. Opo, opossum; tam, tammar; mus, mouse; chk, chicken; zfsh, zebrafish; liz, lizard; ply, platypus.
Discussion
This study is the first to describe IFITM genes in marsupials. Marsupial IFITM genes are similar to those in eutherians with respect to their size (approximately 12 to 17 kDa), structure (two exons that encode two transmembrane domains) and their arrangement within a gene cluster. It also appears that like in mouse and human, there may be multiple transcripts for some of the tammar IFITM genes. Two tammar IFITM genes have a potential ISRE, similar to eutherian IFITMs. The apparent lack of any ISREs in opossum IFITMs is probably an artifact of lack of sufficient 5′ sequence for IFITMA, rather than an actual absence of this motif. ISREs may not be essential for IFITM expression; murine Ifitm3 has two ISREs [15] but mutation of both motifs does not affect the expression of Ifitm3 in the gastrula stage mouse embryo or in fetal gonads [17]. In the tammar several IFITMs are expressed during gastrulation, raising the possibility of a role for IFITM genes in marsupial germ cell development, although localisation of the transcripts is required before any conclusions can be drawn.
Although IFITM genes have been annotated in the genomes of numerous mammalian species (reviewed by [12]), IFITMs from only three mammalian species, the human, mouse and rat, were used for the current comparison. The mouse and human were chosen both because their IFITMs have been well studied and also because these species diverged from each other about 80 million years ago [26], approximately the same length of time as the split between Australasian and South American marsupials [24]. The rat was included to allow comparison of IFITM genes between two closely related species.
Phylogenetic analysis of IFITMs in various vertebrates has been undertaken previously [12] but the current analysis has expanded the data, including all identified marsupial IFITMs and also amphibian IFITMs. In contrast to [12], we also included IFITM10 paralogues. The definition of an IFITM within the literature is vague. All IFITMs contain the human leukocyte antigen cluster of differentiation CD225, yet not all proteins that contain CD225 are IFITMs. The sequences classified as IFITMs by [12] for their analysis included CD225-containing genes that had 2 exons and encoded proteins with a transmembrane domain. These proteins range from 102–157 amino acids in length, with a median length of 132 amino acids. Marsupial, chicken, lizard and rat IFITM10 orthologues all consist of 2 exons and encode proteins 120–130 amino acids long. Mice and humans each have several IFITM10 transcripts, which vary in the number of exons they contain, similar to other IFITM paralogues in these species. Thus, we decided to include IFITM10 paralogues as bona fide IFITM family members.
Our phylogenetic analysis showed that only IFITM5 and IFITM10 have clear orthologues in a range of vertebrate taxa. The orthologous relationships among other IFITM genes are often not very clear. A previous phylogenetic analysis of IFITM genes concluded that there is a high probability of primate-specific gene duplications [12], although concerted evolution cannot be ruled out as an explanation for the higher sequence similarity between paralogues rather than orthologues for many of the IFITMs. The low conservation among IFITM genes, even between species from the same subclass, suggests that either the roles of the different IFITM genes are not conserved between species or, more likely, that there is redundancy between them. The latter idea is strongly supported by an experiment in which the entire IFITM locus was deleted in mice without any apparent effects on normal development [20]. In fact, the study by [20] also suggests that at least during embryonic development, the IFITM family itself may be redundant. Further work is needed to clarify how critical IFITMs are for both embryonic development and general survival. The rapid evolution of IFITMs could be linked to their capacity to act as anti-viral agents; frequent mutation and duplications of these genes may act to counteract virus adaptations [12].
The lack of sequence conservation is mirrored by the lack of conservation of the expression patterns of the different IFITM genes between tammar, mouse and human. Expression data for human and mouse IFITM genes is available from the Unigene expressed sequence tag collection database [27] and some RT-PCR data is also available for the mouse [20]. Even the expression pattern of the most highly conserved IFITM, the tetrapod-specific IFITM10, differs between the three species although expression is more similar between human and tammar than either of these species are to mouse. In mouse Ifitm10 is expressed in the brain and spleen, yet it is absent from these tissues in both the tammar (Figure 4) and human. Conversely, IFITM10 is expressed in bone in tammar and human but not in mouse. The exceptionally high sequence conservation suggests that this gene has an important and conserved function, making it hard to reconcile the apparent lack of conservation in expression patterns. It is also curious that there is almost no information available on this gene in any species: the exception is a brief mention that Ifitm10 ESTs are over-represented in the mouse brain [28].
Examining the expression of IFITM5 in various taxa suggests that both the bone-specific expression of IFITM5 [16, and this study] and the specialised role of IFITM5 in bone production [16] is a recent adaptation specific to therian mammals. It would be interesting to examine IFITM5 expression in tissues from a monotreme mammal but such samples were not available for this study. In zebrafish, in contrast to mammals, IFITM5 is absent from bone, and instead is present in the brain, muscle and liver. The chicken EST database [27] does not include bone but does show high IFITM5 expression in liver, muscle and spleen, with lower levels in the ovary and brain. The expression patterns of IFITM5 in reptiles and amphibians are unknown.
Conclusions
In conclusion, this study has described IFITM gene clusters in two marsupial species and has found evidence to demonstrate that the bone-specific expression of IFITM5 is specific to mammals. It is also the first to recognize the exceptionally high sequence conservation of IFITM10 between different taxa, which suggests an important and conserved (but as yet unidentified) role for IFITM10. This study also suggests that IFITM genes have acquired many different roles during their evolution in vertebrates.
Methods
Bioinformatics
IFITMs in the opossum genome were identified using the BLAT search engine on the UCSC Genome Bioinformatics website [29]. The region including and surrounding opossum IFITM5 (chrUn:15587203–16132202, Oct. 2006 (Broad/monDom5) incorporating ~545 kb of sequence) was searched for predicted genes using GENSCAN [30]. Paralogues of IFITM5 were identified by using Ensembl [31]. Primers based on human or opossum IFITMs were used to amplify tammar IFITMs by RT-PCR. The resulting products were cloned and sequenced and these sequences were then used to search the M. eugenii whole genomic shotgun (WGS) trace archives using BLASTn and tBLASTn [32] with discontiguous megablasts. This usually yielded small contigs containing either the first or second exon of each gene. These contigs were assembled using the online CAP3 program [33]. Sequence from these was then used to design tammar-specific primers within the 5 and 3 prime UTRs of each gene (Table 3). The ORF of each tammar IFITM gene was amplified by RT-PCR, cloned and sequenced.
Characterisation of the tammar IFITM gene cluster and FISH
To characterise the tammar IFITM gene cluster, a male tammar genomic BAC library obtained from Arizona Genomics Institute (Tucson, AZ, USA) was screened using 32P-labelled probes as described previously [34]. Probes to tammar IFITMA2 and IFITM10 were labelled using the Megaprime DNA labelling kit (GE Healthcare, NSW, Aust.). DNA was extracted from the resulting positive BAC clones using the PhasePrep BAC DNA kit (Sigma-Aldrich, NSW, Aust.) and the purified DNA was shotgun sequenced at the Australian Genome Research Facility (Qld, Aust.).
Fluorescence in situ hybridisation (FISH) was performed as described previously [35]. Briefly, the purified BAC genomic clones were labelled with dUTP-digoxygenin (DIG) by nick translation at 14°C for about one hour using the Megaprime DNA Labelling kit (GE Healthcare, Aust.) and then co-precipitated with tammar Cot-1 DNA. Tammar metaphase chromosomes from testis were then incubated with the labelled probes overnight at 37°C. Bound probe was detected using a mouse anti-DIG FITC–labelled antibody (Roche, NSW, Aust.) and chromosomes were counterstained with DAPI (4, 6-diamidino- 2-phenylindole).2
Protein alignments and phylogenetic tree construction
Amino acid sequences of mouse, human and rat IFITMs were obtained from the National Centre for Biotechnology Information [32]. Genomes of the platypus, chicken, lizard, frog, fish, lamprey, sea squirt, lancelet, sea urchin, mollusc, fly and nematode were searched for IFITM homologues (Additional file 1: Table S 1) using the UCSC Genome Bioinformatics [29], Ensembl [31] and NCBI websites [32]. Multisequence amino acid alignments were performed using ClustalW [36] and edited with GeneDoc [37]. A phylogenetic tree of vertebrate IFITMs for which full length (or almost full length) sequence was available was constructed using Phylip [38]. First, multi-sequence alignment of amino acids was performed using ClustalW using the PAM weight matrix. Then, in Phylip, the PAM distance matrix was calculated in Protdist. The matrix was transformed into a Neighbour joining tree and then a majority-rule consensus tree for 1000 bootstraps was drawn using Consense.
Expression analysis by RT-PCR
Tissues were obtained from tammars in our University of Melbourne breeding colony. Samples were collected from females carrying embryos or fetuses as described previously [39, 40]. Briefly, for the pre-natal stages, embryos up to and including the early somite stage were frozen whole. Mid-somitogenesis stage embryos with their adjoining vascular (trilaminar) yolk sac were separated from their avascular (bilaminar) yolk sac and the two regions were frozen separately. All later stages were divided into embryo or fetus, vascular yolk sac placenta and avascular yolk sac placenta and each region was frozen separately. Total RNA was extracted from adult tissues using Tri Reagent (Ambion Inc, Texas CA., USA) and from conceptuses using the GenElute Mammalian Total RNA Miniprep kit (Sigma Aldrich, NSW, Aust.) according to kit protocols. RNA was DNAse-treated with DNA-Free (Ambion) and 40 ng of RNA was reverse-transcribed using SuperScript III (Invitrogen, CA, USA) in a total volume of 20 μL. PCR was performed using GoTaq (Promega, NSW, Aust.) in a 30 μL reaction, which included 0.5 μL of cDNA and primers at a final concentration of 0.5 μM. PCR amplification involved an initial 2 minute denaturation step at 94°C and extension at 72°C for 30 seconds. The number of amplification cycles and the annealing temperatures for each gene are listed in Table 3. All experiments were approved by the University of Melbourne Animal Experimentation Ethics Committees and all animal handling and husbandry were in accordance with the National Health and Medical Research Council of Australia (2004) guidelines.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- BLAST:
-
Basic local alignment search tool
- BYS:
-
Bilaminar yolk sac
- CD225:
-
Cluster of differentiation antigen
- EST:
-
Expressed sequence tag
- FISH:
-
Fluorescence in situ hybridisation
- IFITM:
-
Interferon inducible transmembrane protein
- ISRE:
-
Interferon-stimulated response element
- NCBI:
-
National Centre for Biotechnology Information
- ORF:
-
Open reading frame
- PGCs:
-
Primordial germ cells
- shRNA:
-
Short hairpin RNA
- UTR:
-
Untranslated region
- WGS:
-
Whole genomic shotgun.
References
Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR: Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 1984, 38: 745-755. 10.1016/0092-8674(84)90270-8.
Reid LE, Brasnett AH, Gilbert CS, Porter ACG, Gewert DR, Stark GR, Kerr IM: A single DNA response element can confer inducibility by both α- and γ-interferons. Proc Nat Acad Sci USA. 1989, 86: 840-844. 10.1073/pnas.86.3.840.
Lewin AR, Reid LE, McMahon M, Stark GR, Kerr IM: Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991, 199: 417-423. 10.1111/j.1432-1033.1991.tb16139.x.
Evans SS, Lee DB, Han T, Tomasi TB, Evans RL: Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood. 1990, 76: 2583-2593.
Evans SS, Collea RP, Leasure JA, Le DB: IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J Immunol. 1993, 150: 736-747.
Delbrandre GA, Marinx OP, Evans SS, Majjaj S, Leo O, Caput D, Huez GA, Wathelet MG: Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J Biol Chem. 1995, 270: 23860-23866. 10.1074/jbc.270.40.23860.
Brem R, Oraszlan-Szovik K, Foser S, Bohrmann B, Certa U: Inhibition of proliferation by 1-8U in interferon-alpha-responsive and nonresponsive cell lines. Cell Mol Life Sci. 2003, 60: 235-1248.
Daniel-Carmi V, Makovitzki-Avraham E, Reuven EM, Goldstein I, Zilkha N, Rotter V, Tzehoval E, Eisenbach L: The human 1-8D gene (IFITM2) is a novel p53 independent pro-apopototic gene. Int J Cancer. 2009, 125: 2810-2819. 10.1002/ijc.24669.
Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ: IFITM proteins mediate the innate immune response to influenza A H1N1 virus, West Nile virus and Dengue virus. Cell. 2009, 139: 1243-1254. 10.1016/j.cell.2009.12.017.
Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M: Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and Influenza A virus. PLoS Pathology. 2011, 7: e1001258-10.1371/journal.ppat.1001258.
Lu J, Pan Q, Rong L, He W, Liu SL, Liang C: The IFITM proteins inhibit HIV-1 infection. J Virol. 2011, 85: 2126-2137. 10.1128/JVI.01531-10.
Siegrist F, Ebeling M, Certa U: The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res. 2011, 31: 183-197. 10.1089/jir.2010.0112.
Saitou M, Barton SC, Surani MA: A molecular programme for the specification of germ cell fate. Nature. 2002, 418: 293-300. 10.1038/nature00927.
Tanaka SS, Matsui Y: Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev. 2002, 119 (Suppl 1): S261-S267.
Lange UC, Saitou M, Western PS, Barton SC, Surani MA: The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003, 3: 1-10.1186/1471-213X-3-1.
Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, Godin E, de Tambasco Oliveira P, Atkins GJ, Nanci A, Thomas G: Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008, 23: 1497-1508. 10.1359/jbmr.080412.
Tanaka SS, Yasuka L, Yamaguchi YL, Tsoi B, Lickert H, Tam PP: IFITM/Mil/Fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Developmental Cell. 2005, 9: 745-756. 10.1016/j.devcel.2005.10.010.
Lickert B, Cox B, Wehrle C, Taketo MM, Kemler R, Rossant J: Dissecting Wnt/β-catenin signaling during gastrulation using RNA interference in mouse embryos. Development. 2005, 132: 2599-2609. 10.1242/dev.01842.
Hanagata N, Li X, Morita H, Takemura T, Li J, Minowa T: Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J Bone Miner Metab. 2010, 29: 279-290.
Lange UC, Adams DJ, Lee C, Barton S, Schneider R, Bradley A, Surani MA: Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Molec Cell Biol. 2008, 28: 4688-4696. 10.1128/MCB.00272-08.
Morel N, Brocher G, Synguelakis M, Le Gal La Salle G: Immunological identification of a new 14 × 103 Mr membrane bound protein in Torpedo electric organ. J Cell Sci. 1991, 98: 351-361.
Johnson MC, Sangrador-Vegas A, Smith TJ, Cairns MT: Cloning and characterization of two genes encoding rainbow trout homologues of the IFITM protein family. Vet Immunol Immunopathol. 2006, 110: 357-362. 10.1016/j.vetimm.2005.12.007.
Luo ZX, Yuan CX, Meng QJ, Ji Q: A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011, 476: 442-445. 10.1038/nature10291.
Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A: The delayed rise of present-day mammals. Nature. 2007, 446 (7135): 507-512. 10.1038/nature05634.
Renfree MB, Papenfuss AT, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G, Wong ES, Lefèvre CM, Nicholas KR, Kuroki Y, Wakefield MJ, Zenger KR, Wang C, Ferguson-Smith M, Nicholas FW, Hickford D, Yu H, Short KR, Siddle HV, Frankenberg SR, Chew KY, Menzies BR, Stringer JM, Suzuki S, Hore TA, Delbridge ML, Patel H, Mohammadi A, Frankenberg SR, Schneider NY, Hu Y, O'Hara W, Al Nadaf S, Wu C, Feng ZP, Cocks BG, Wang J, Flicek P, Searle SM, Fairley S, Beal K, Herrero J, Carone DM, Suzuki Y, Sugano S, Toyoda A, Sakaki Y, Kondo S, Nishida Y, Tatsumoto S, Mandiou I, Hsu A, McColl KA, Lansdell B, Weinstock G, Kuczek E, McGrath A, Wilson P, Men A, Hazar-Rethinam M, Hall A, Davis J, Wood D, Williams S, Sundaravadanam Y, Muzny DM, Jhangiani SN, Lewis LR, Morgan MB, Okwuonu GO, Ruiz SJ, Santibanez J, Nazareth L, Cree A, Fowler G, Kovar CL, Dinh HH, Joshi V, Jing C, Lara F, Thornton R, Chen L, Deng J, Liu Y, Shen JY, Song XZ, Edson J, Troon C, Thomas D, Stephens A, Yapa L, Levchenko T, Gibbs RA, Cooper DW, Speed TP, Fujiyama A, M Graves JA, O'Neill RJ, Pask AJ, Forrest SM, Worley KC: Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011, 12: R81-10.1186/gb-2011-12-8-r81. [http://bioinf.wehi.edu.au/tammar]
Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS: Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001, 294: 2348-2351. 10.1126/science.1067179.
Schuler GD: Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J Mol Med. 1997, 75: 694-698. 10.1007/s001090050155. [http://www.ncbi.nlm.nih.gov/UniGene/]
Plessy C, Fagiolini M, Wagatsuma A, Harasawa N, Kuji T, Asaka-Oba A, Kanzaki Y, Fujishima S, Waki K, Nakahara H, Hensch TK, Carninci P: A resource for transcriptomic analysis in the mouse brain. PLoS One. 2008, 3: e3012-10.1371/journal.pone.0003012.
Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ: The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011, 39: D871-875. 10.1093/nar/gkq1017. [http://www.genome.ucsc.edu/]
Burge C, Karlin S: Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997, 268: 78-94. 10.1006/jmbi.1997.0951. [http://genes.mit.edu/GENSCAN.html]
Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Overduin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E: The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2007, 35: D610-D617. 10.1093/nar/gkl996. [http://www.ensembl.org/index.html]
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol. 1990, 215: 403-410. [http://www.ncbi.nlm.nih.gov]
Huang X, Madan A: CAP3: A DNA sequence assembly program. Genome Res. 1999, 9: 868-877. 10.1101/gr.9.9.868. [http://pbil.univ-lyon1.fr/cap3.php]
Deakin JE, Koina E, Waters PD, Doherty R, Patel VS, Delbridge ML, Dobson B, Fong J, Hu Y, van den Hurk C, Pask AJ, Shaw G, Smith C, Thompson K, Wakefield MJ, Yu H, Renfree MB, Graves JA: Physical map of two tammar wallaby chromosomes: A strategy for mapping in non-model mammals. Chrom Res. 2008, 16: 1159-1175. 10.1007/s10577-008-1266-y.
Yu H, Pask AJ, Shaw G, Renfree MB: Comparative analysis of the mammalian WNT4 promoter. BMC Genomics. 2009, 10: 416-10.1186/1471-2164-10-416.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG: Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23: 2947-2948. 10.1093/bioinformatics/btm404. [http://www.ebi.ac.uk/Tools/msa/clustalw2/]
Nicholas KB, Nicholas HB Jr, Deerfield DW II: GeneDoc: Analysis and visualization of genetic variation. EMBNEW.NEWS. 1997, 4: 14-[http://www.nrbsc.org/gfx/genedoc/]
Felsenstein J: PHYLIP-Phylogeny Inference Package (Version 3.2). Cladistics. 1989, 5: 164-166. [http://www.phylip.com/]
Renfree MB, Tyndale-Biscoe CH: Manipulation of marsupial embryos and pouch young. Methods of Mammalian Reproduction. Edited by: Daniel JC. 1978, Academic, NewYork, 307-331.
Hickford D, Frankenberg S, Renfree MB: The tammar wallaby, Macropus eugenii: a model kangaroo for the study of developmental and reproductive biology. Emerging Model Organisms: A Laboratory Manual. Volume 2. Edited by: Crotty DA, Gann A. 2010, Cold Spring Harbor Laboratory Press, New York, 449-494.
Acknowledgments
We thank all members of the wallaby research group, particularly Bonnie Dopheide for assistance with FISH, Hongshi Yu for assistance with screening the BAC library and Scott Brownlees and Kerry Martin for their help with the animals. DH was supported by an ARC Postdoctoral Fellowship, MBR by an Australian Research Council Federation Fellowship. Funding for the project was provided by an ARC Discovery grant to MBR and GS and by the ARC Centre of Excellence for Kangaroo Genomics.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DH carried out the experimental work and participated in the bioinformatic analyses and drafted the manuscript. SF participated in the bioinformatic analyses. GS and MBR collected embryo specimens and SF, GS and MBR revised the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hickford, D., Frankenberg, S., Shaw, G. et al. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics 13, 155 (2012). https://doi.org/10.1186/1471-2164-13-155
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-13-155