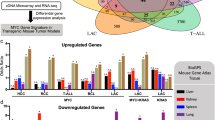
Abstract
Background
The transcription factor MYC is a critical regulator of diverse cellular processes, including both replication and apoptosis. Differences in MYC-regulated gene expression responsible for such opposing outcomes in vivo remain obscure. To address this we have examined time-dependent changes in global gene expression in two transgenic mouse models in which MYC activation, in either skin suprabasal keratinocytes or pancreatic islet β-cells, promotes tissue expansion or involution, respectively.
Results
Consistent with observed phenotypes, expression of cell cycle genes is increased in both models (albeit enriched in β-cells), as are those involved in cell growth and metabolism, while expression of genes involved in cell differentiation is down-regulated. However, in β-cells, which unlike suprabasal keratinocytes undergo prominent apoptosis from 24 hours, there is up-regulation of genes associated with DNA-damage response and intrinsic apoptotic pathways, including Atr, Arf, Bax and Cycs. In striking contrast, this is not the case for suprabasal keratinocytes, where pro-apoptotic genes such as Noxa are down-regulated and key anti-apoptotic pathways (such as Igf1-Akt) and those promoting angiogenesis are up-regulated. Moreover, dramatic up-regulation of steroid hormone-regulated Kallikrein serine protease family members in suprabasal keratinocytes alone could further enhance local Igf1 actions, such as through proteolysis of Igf1 binding proteins.
Conclusions
Activation of MYC causes cell growth, loss of differentiation and cell cycle entry in both β-cells and suprabasal keratinocytes in vivo. Apoptosis, which is confined to β-cells, may involve a combination of a DNA-damage response and downstream activation of pro-apoptotic signalling pathways, including Cdc2a and p19Arf/p53, and downstream targets. Conversely, avoidance of apoptosis in suprabasal keratinocytes may result primarily from the activation of key anti-apoptotic signalling pathways, particularly Igf1-Akt, and induction of an angiogenic response, though intrinsic resistance to induction of p19Arf by MYC in suprabasal keratinocytes may contribute.
Similar content being viewed by others
Background
The c-MYC proto-oncogene encodes a transcription factor, c-MYC (MYC), which regulates the expression of cellular targets involved in a wide range of diverse cellular functions, including cell growth, proliferation, loss of cell-cell contact, loss of differentiation and angiogenesis [1–8]. While the predominant role of physiological MYC in most tissues is to promote G1/S transition in the cell cycle (and thereby proliferation) [1, 9, 10] and inhibit differentiation [11–13], deregulated MYC (oncogenic) can lead to uncontrolled proliferation and tumour growth [3]. Paradoxically though, MYC is able to act as its own tumour suppressor, as deregulated MYC activity can also promote apoptosis (both in vitro[14–16] and in vivo[17]) and senescence [18, 19]. See [20] for a recent review of the MYC field. Such linkage between seemingly opposing functions - proliferation and apoptosis - is also found in other cell-cycle-associated genes, such as E2f, E1a and c-Fos[21].
The mechanisms by which MYC elicits the vast host of biological responses for which it appears to be responsible are not yet fully understood. Currently, around 1,700 genes have been classified as putative MYC targets [22, 23] using methods such as serial analysis of gene expression (SAGE) [24], DNA microarrays [3] and subtractive hybridization [25]. It has been hypothesized that MYC may have the potential to regulate up to 15% of the entire genome [26], leading to it being described as a 'master regulator' of gene expression.
Regulatable transgenic mouse models have allowed controlled activation of a modified MYC-containing chimaeric transcription factor (MYC-ERTAM) in distinct cell populations in adult mice, such as the pancreatic islet β-cells [27] and suprabasal keratinocytes (SBK) of skin epidermis [28]. Our previous work has shown that continuous activation of MYC-ERTAM in these diverse tissues exposes the dual potential of MYC to activate pathways involved in cell replication and cell death under differing environmental conditions. In suprabasal epidermis, MYC promotes entry of post-mitotic keratinocytes into the cell cycle, concomitant with loss of differentiation and increased vascularization leading to formation of pre-cancerous papillomas [28]. In contrast, although MYC promotes rapid cell cycle entry of pancreatic β-cells, these cells quickly proceed to undergo apoptosis leading to severe cell depletion and diabetes [27]. This indicates a crucial role for tissue context and the surrounding micro-environment in determining cell fate.
The divergence of MYC-induced phenotypes between these two tissues has enabled us to compare MYC-regulated gene expression patterns over a time course of MYC-ERTAM activation, by employing high-throughput transcriptome analysis using microarrays. Comparison of the transcriptional response between the two tissues identified potential signalling pathways which may promote apoptosis of β-cells and prevent apoptosis in SBK; the DNA-damage response pathway, and the Insulin-like growth factor 1 (Igf1) signalling pathway, respectively. In addition, up-regulation of angiogenesis-related genes and of those encoding members of the steroid hormone-regulated Kallikrein serine protease family was found in SBK but not in β-cells. Kallikreins may increase availability and action of Igf1 through proteolysis of Igf1 binding proteins. Together with angiogenesis, Kallikreins may provide a local tissue-specific regulatory mechanism for determining ultimate MYC function in vivo.
Results and Discussion
Activation of MYC-ERTAM mediates transcription of genes involved in a wide range of cellular functions
Time courses were set up following activation of MYC in β-cells (pancreas) and SBK (skin) via administration of 4-hydroxytamoxifen (4OHT) for 4 hrs, 8 hrs, 16 hrs and 32 hrs as described (Methods). Vehicle-treated samples acted as direct time point controls for 4OHT-treated samples. Laser capture microdissection (LCM) was utilized to allow isolation of pancreatic islet tissue. Significant gene expression changes for the main experimental conditions (4OHT treatment, tissue type and time point following initial treatment) and their interactions, as well as information on the effects of further covariates such as batch effects and RNA quality, were identified using a custom R package, Envisage (available on request).
Analysis of gene expression for 12,349 curated probe sets (Methods) identified 6,633 unique genes as being significantly altered following activation of MYC with a false discovery rate of 5%. 1,615 genes showed significant effects for the joint effects of 4OHT treatment, time and tissue type; 2,015 genes showed significant effects for the interaction between 4OHT treatment and tissue type; 2,221 genes showed significant effects for the interaction between 4OHT treatment and time; and 1,843 genes showed significant effects for the main effect of 4OHT treatment only. Of the MYC-responsive genes, the expression levels of 1,199 were altered greater than 2-fold (up- or down-regulated) after only 4 hours of 4OHT treatment for the pancreatic β-cells, while only 530 were similarly affected for SBK. However, at 8 hours following initial 4OHT treatment, the expression levels of 1,905 and 1,882 were altered greater than 2-fold for the pancreas and the skin respectively (Figure 1A). This suggests a more prominent initial response to MYC in β-cells compared to SBKs.
Transcriptional response to MYC-ERTAM activation. A) Activation of MYC-ERTAM led to a significant change in expression of a large number of genes within only 4 hours for the pancreas (≥ 2-fold change, p ≤ 0.05). A similar transcriptional response was not detected until 8 hours post-MYC-ERTAM activation for SBK. B) Analysis of 6,633 genes, whose expression was altered within 8 hours upon activation of MYC-ERTAM in either pancreas or skin (green bars), identified enrichment of genes in a wide range of MYC-mediated functions. Analyses of genes whose expression was altered greater than 2-fold within 8 hours for the SBK (red bars) and the pancreatic β-cells (blue bars) showed a similar level of response to MYC-ERTAM activation in both tissues within 8 hours, with a slight increase in the level of response for the pancreas as compared to the skin, in particular for DNA-replication and response to DNA-damage. p-values represent significance of enrichment of GO classes for the 6,633 genes. C) Genes identified as showing significant differential expression due to the joint effects of MYC-ERTAM activation and tissue (with or without an interaction with the time point) were clustered to identify functionally related genes (red, up-regulated; black, no change; green, down-regulated). The blow-up identifies a group of functionally related genes showing increased expression in SBK but not islet β-cells.
Gene ontology (GO) enrichment analysis using the DAVID functional annotation tool [29] identified enrichment of genes involved in myriad cellular functions following activation of MYC. The majority of MYC-responsive genes were involved in metabolic, transcriptional, transportational and signal transduction pathways (Figure 1B; green bars). Genes involved in post-transcriptional modification (e.g. RNA processing, RNA splicing) and post-translational modification (e.g. protein catabolic process, protein folding, protein localization and ubiquitin-dependent catabolic process) were also significantly enriched. Similarly, a significant enrichment of genes relating to ribosome biogenesis was detected, suggestive of MYC's recently elucidated role as a regulator of ribosome biogenesis and protein synthesis [30]. As expected given the role of MYC in proliferation, genes involved in cell cycle progression were amongst the most significantly enriched (p = 1.21 × 10-17). Genes involved in apoptosis and DNA-damage checkpoint pathways were also enriched (p = 7.81 × 10-3 and p = 5.07 × 10-5 respectively), along with genes involved in related functions such as cellular response to stress (p = 1.56 × 10-8) and cytoskeleton organisation (p = 9.12 × 10-5).
Enrichment of GO terms for MYC-responsive genes showing early changes in expression (≥ 2-fold change within the first 8 hours of MYC activation) for the pancreas (Figure 1B; blue bars) and skin (Figure 1B; red bars) identified similar numbers for both tissues, with the exception of genes relating to DNA-damage and DNA replication, where a larger number of genes are detected for the pancreas than for the skin. These results indicate that whilst expression of genes relating to multiple cellular functions is common to both tissues, expression of genes involved in DNA-damage and replication is more specific to the β-cells.
Expression of putative MYC target genes following MYC-ERTAM activation
The MYC Target Gene Database [31] currently identifies 1,697 genes as putative MYC targets [22]. Of these, 13.4% and 19.2% were found to be both MYC-responsive and show a 2-fold change in expression in the skin and pancreas respectively within 8 hours (Additional file 1, Table S1). The predominant role for these genes was in DNA replication, biosynthesis, metabolism, cell cycle, cell division and other related functions. Cellular functions relating to apoptosis and cell death were also highly enriched, although to a lesser degree than those relating to cellular proliferation. These data suggest that activation of the MYC-ERTAM protein in vivo leads to a rapid change in the expression of a large number of putative MYC targets. However, known target genes represent only a small fraction of detected MYC-responsive genes, indicating that the majority of these observed expression changes may be downstream of direct MYC-induced transcription.
To identify the level of overlap between the genes classed as significantly altered in this study and those identified in previous analyses, we utilised the Gene Set Enrichment Analysis (GSEA) program developed by the Broad Institute [32]. This allowed us to identify gene sets in which significant differentially expressed genes are enriched. Gene sets were taken from the Molecular Signatures Database (MSigDB), as well as from additional published datasets. Additional file 1, Table S7 and Additional file 1, Table S8 show the results from GSEA (FDR < 0.01) for the genes showing significant expression at the early time points (4 hours and 8 hours) for the pancreas and skin, respectively. These results indicate a clear enrichment in the pancreas of genes annotated as being related to MYC function from previous publications, including MYC target genes from the MYC Target Gene Database [22], genes indicative of a MYC-induced oncogenic signature from Blid et al.[33] and genes up-regulated in the studies of Coller et al., Schumacher et al., Yu et al., and Lee et al.[3, 34–36]. Similarly, down-regulated genes showed enrichment with down-regulated genes from the study of Yu et al.[36]. In comparison, genes up-regulated in the skin are enriched for genes also found to be up-regulated in the studies of Coller et al., Yu et al. and Zeller et al.[3, 22, 36], while MYC target genes from the MYC Target Gene Database [22], genes indicative of a MYC-induced oncogenic signature from Blid et al.[33] were enriched with an FDR value < 0.017. This suggests that the gene expression signature identified in both the pancreas and skin is indicative of changes in expression related to MYC function.
Cell cycle response following MYC activation
One of the key functions of the MYC onco-protein is promotion of cell cycle progression, particularly G1/S phase transition [1, 2, 37]. Activation of MYC in suprabasal keratinocytes (SBK) and pancreatic β-cells has been previously shown to initiate G1/S transition in target cells [27, 28, 38], and this was seen through immunohistological staining with anti-Ki67 antibodies (Figure 2A), as well as in the response detected in genes relating to cell cycle progression by gene ontology classification (Figure 1B and Additional file 1 Table S2). A subset of some interesting genes from this list is shown in Table 1.
Cell cycle following activation of MYC-ERTAM. A) Sections from 4OHT-treated MYC-ERTAM transgenic mice were stained with Ki67-specific antibodies (green) and a tissue specific antibody (red; insulin for β-cells, keratin 1 for SBK). DAPI staining (blue) identifies cell nuclei. No Ki67 staining was detected in β-cells or SBK in vehicle-treated samples, although proliferating basal cells in the skin showed clear staining. Following 32 hours of 4OHT, staining of both β-cells and SBK was detected, indicated by concomitant staining of Ki67 and the tissue-specific protein. Images shown are representative sections from different levels through replicate tissue samples. b, basal keratinocyte; sb, suprabasal keratinocyte. B) The change in expression of genes relating to cell cycle following activation of MYC-ERTAM is represented as a heatmap (red, up-regulated; black, no change; green, down-regulated). Significant change in expression is detected for key early G1-S phase transition genes such as Ccnd1, Ccnd2 and Ccne2, indicating activation of cell cycle progression, particularly for the pancreas. C) qRT-PCR validation for gene expression changes in genes relating to cell cycle progression indicated a clear proliferative response in the pancreatic β-cells following MYC-ERTAM activation. In particular, genes relating to G1/S-phase transition such as Ccnd1, Ccnd2 and Ccne2 showed early changes in expression (8 hours), whilst genes whose products relate to later cell cycle events such as Ccna2 and Ccnb1 showed increased expression at later time points (32 hours). Few significant changes were detected for these key proliferation markers in SBK. This may be due to the relatively low proportion of keratinocytes that are responsive to Myc-induced proliferation at these early time-points, whilst differentiated keratinocytes of the granular layer are refractory to the proliferative influence of MYC.
Activation of MYC in the β-cells resulted in a change in expression of 213 cell cycle- and proliferation-related genes within 8 hours of MYC activation, with 116 genes up-regulated and 101 genes down-regulated (Additional file 1, Table S2). Pcna, a MYC target gene associated with the cell cycle, was expressed in both pancreatic β-cells and SBK throughout most of the time-course, with Cdc25a expressed only in the former. In β-cells, cyclin genes Ccnd1, Ccnd2 and Ccne2, whose products are necessary for G1/S-phase transition in the cell cycle, were up-regulated within 4 hours of MYC activation. Cyclin genes Ccna2, Ccnb1 and Ccne1, whose products are involved in later G1/S-phase and G2/M-phase cell cycle events, were up-regulated greater than 3-fold subsequently at 8 hours (Table 1 and Figure 2C).
Activation of MYC in the SBK resulted in a less prominent cell cycle response compared to β-cells, with a change in expression of 144 cell cycle- and proliferation-related transcripts within 8 hours of MYC activation - 73 genes up-regulated and 74 genes down-regulated (Additional file 1, Table S2). Of G1/S-phase cell cycle genes, Ccnd2 and Ccnd3 showed a ≥ 2-fold increase in expression at 8 hours. In contrast to β-cells, later cell cycle genes such as Ccna2 and Ccne2 were either down-regulated or unchanged in SBK. Interestingly, the Ccnb1 gene whose product, cyclin B1, is involved predominantly in later cell cycle events (G2/M transition), was down-regulated 2-fold in SBK early in the time-course. Cyclin B1 interacts with Cdc2a (Cdk1) to form the mitotic initiation complex, which was also down-regulated by ~2-fold at 4 hrs in epidermis. This contrasts with β-cells, in which both Ccnb1 and Cdc2a showed significant up-regulation. Although speculation here, these findings may suggest SBK are able to enter the G1/S phase of the cell cycle, which is a known function of MYC, but not progress through the G2/M phase. Interestingly, it was previously shown that over-expression of MYC causes a P53-dependent G2 arrest in normal fibroblasts [39]. Such cells may then be enforced by MYC to reinitiate DNA replication, resulting in aneuploidy.
Of these cell cycle-related genes, Ccna2[40] and Ccnd1[41, 42] have been previously designated as putative MYC targets through high-throughput screening, and Ccnb1[43] and Ccnd2[44–46] have been previously confirmed as direct transcriptional targets of MYC through the use of chromatin Immunoprecipitation (ChIP) analysis. The cyclin D2-related kinase Cdk4, also a previously characterized direct MYC target [47], showed increased expression after 4 hours of MYC activation in the pancreas, with a 6-fold increase detected subsequently at 16 hours. Cdk4 was also found to be highly up-regulated at 8 hours in the skin, with a fold change of almost 12 (Table 1). No significant change was detected for the cyclin E-associated CDK gene Cdk2 in either the skin or the pancreas, However Cdk7, which has a role in both activating cyclin complexes and regulating transcription, was up-regulated at 8 hours in the skin.
Down-regulation of another known MYC target gene, the cyclin dependent kinase (CDK) inhibitor Cdkn1b (p27Kip1), which inhibits G1/S-phase transition by association with the cyclin E-Cdk2 complex [48], was detected for both the skin and the pancreas. Also, the expression of Cks2, a MYC target gene whose product is involved in degradation of p27Kip1, increased from 8 hours following MYC activation in the pancreas. Interestingly, the Cdc2a gene, whose product Cdk1 is essential for mammalian cell division [49, 50], was also found to be highly up-regulated in β-cells (Table 1). Cdk1 has been found to substitute for other CDKs to drive cell cycle progression [51], and is particularly associated with Cdk4 in G1/S-phase progression [reviewed in 52]. This may indicate a significant role for Cdk1 in the promotion of cell cycle progression following MYC activation in β-cells. Alternatively, it has been shown that premature activation of Cdk1 can lead to mitotic catastrophe in G2/M-phase and apoptosis (upstream of p53-induced mitochondrial outer membrane permeabilization) in neurons [53]. Given that this CDK was also detected at later time points, this may indicate a possible role for Cdk1 in the MYC-induced apoptosis pathways. In addition to this, the CDK inhibitor Cdkn2c (p18Ink4c), which inhibits G1/S-phase transition via interactions with Cdk4 [54] and Cdk6 [55], was down-regulated early in the pancreas. However, by 16 hours expression levels had risen dramatically, which may be indicative of cell cycle arrest prior to apoptosis. In addition, the CDK inhibitor Cdkn1a (p21Cip1) - a downstream target of the tumour suppressor p53 - was up-regulated at 8 hours (Table 1).
Growth arrest specific gene 6 (Gas6) was up-regulated in SBK (Table 1), and has been found to have different roles dependant on tissue location, including promotion of cell cycle progression in fibroblasts and Schwann cells [56, 57], and has been shown to play a role in cellular survival by inducing signalling through the PI3K/Akt pathway leading to phosphorylation of the forkhead box protein Foxo1 in endothelial cells [58] and in oligodendrocytes of the central nervous system [59]. Interestingly, the growth arrest and DNA-damage-inducible 45 gamma gene, Gadd45g, a proposed MYC target whose product is involved in growth arrest at the G2/M DNA-damage checkpoint [60], showed increased expression at 4 hours in SBK, and remained 3-fold up-regulated throughout the time course, whilst down-regulation at 8 hours was detected in β-cells. This suggests potential activation of pathways to limit unchecked proliferation in the keratinocytes (Table 1).
Genes relating to increased cellular mass, cytoskeleton organization and DNA replication were also detected for SBK (Additional file 1, Table S3), including the membrane skeletal proteins Adducin 1 (Add1) and Pdlim3, the actin-modulating protein Cofilin 1 (Cfl1), members of the kinesin family of microtubule motor proteins, members of the myosin superfamily of actin binding motor proteins (e.g. Myh4, Myoc, Myo1f), and members of the tubulin family of microtubule proteins (Tuba3a, Tubb2c, Tubb6). Plectin 1 (Plec1), one of the main components of the cytoskeleton, showed an increase in expression of roughly 3- to 4-fold throughout the early stages of the time course (Table 1). This increased activity of microtubule formation and actin formation for both the pancreas and skin is indicative of increased cellular turnover in both tissues.
Apoptotic response following MYC activation
The ultimate phenotypic response to activation of MYC in pancreatic β-cells is apoptosis [27]. Immunohistological staining for Caspase 3, an early marker for initiation of apoptosis pathways, indicated an apoptotic response to MYC activation in the β-cells but not in the SBK (Figure 3A). In contrast, MYC-activated SBK that have begun a process of terminal differentiation, re-enter the cell cycle but are protected from conventional apoptosis [33, 38]. These cells will ultimately be shed and removed from the surrounding micro-environment thus ridding the host of potentially harmful pre-cancerous cells [61]. Our array data confirm a large transcriptional response detected in genes relating to apoptosis and survival by gene ontology classification in both tissues (Figure 3B and Additional file 1 Table S4). A subset of important genes from this list is shown in Table 2.
Apoptosis following activation of MYC-ERTAM. A) Sections from 4OHT-treated MYC-ERTAM transgenic mice were stained with Caspase 3-specific antibodies (green) and a tissue specific antibody (red; insulin for β-cells, keratin 1 for SBK). DAPI staining (blue) identifies cell nuclei. No Caspase 3 staining was detected in β-cells or SBK in vehicle-treated (VT) samples. Following 32 hours of 4OHT, Caspase 3 was detected in β-cells, but no Caspase 3 activity was detected in treated SBK indicating that apoptosis is specific to the β-cell model. Images shown are representative sections from different levels through replicate tissue samples. b, basal keratinocyte; sb, suprabasal keratinocyte. B) The change in expression of genes relating to apoptosis and cell survival following activation of MYC-ERTAM is represented as a heatmap (red, up-regulated; black, no change; green, down-regulated). C) qRT-PCR validation for gene expression changes in genes relating to apoptosis and cell survival confirms disparate changes in gene expression between skin and pancreas. Genes relating to DNA-damage in particular, such as Atr and Chk2, show a clear increase in expression for pancreatic β-cells, which confirms changes seen in microarray data, as did apoptosis genes Cycs and Endog. As with the microarray data, up-regulation of the Igf1 gene was detected from 8 hours for the skin following MYC-ERTAM activation, with an increase in expression also detected for the pancreas at the later 32 hour time point.
Activation of MYC in pancreatic β-cells identified a significant change in expression for 92 genes relating to cell death and apoptosis. Of these, 42 genes showed an increase and 50 genes showed a decrease in expression (Additional file 1, Table S4). Early activation of key regulators of apoptosis featured prominently in these data. Activation of MYC in SBK resulted in significant changes in expression for 66 genes relating to apoptosis and cell survival, including 37 genes showing an increase and 29 genes showing a decrease in expression (Additional file 1, Table S4).
The tumour suppressor Cdkn2a, which encodes for the CDK inhibitor p16Ink4a and the alternative reading frame tumour suppressor p19Arf (p14Arf in humans), was up-regulated 2-fold at 4 hours and remained at an elevated level throughout the time course for β-cells, but remained unchanged in SBK (Table 2 and Figure 3C). Previous studies have identified a possible role for the p19Arf/p53/Mdm2 tumour suppressor pathway in MYC-mediated apoptosis [62, 63]. Importantly, data from the Evan lab has elegantly shown that high levels of MYC-ERTAM activation present in this transgenic model, led to expression of p19Arf concomitant with apoptosis [64]. In contrast, low levels of MYC-ERTAM activation did not induce apoptosis or p19Arf but rather β-cell proliferation. However, Finch et al. previously showed that loss of p19Arf in β-cells of the MYC-ERTAM transgenic model resulted in mainly increased proliferation, not suppression of apoptosis [65]. Thus the role of p19Arf in defending against aberrant oncogenic MYC-induced hyper-proliferation may be related to cell cycle arrest, and not directly to apoptosis pathways in β-cells.
The anti-apoptotic function of BclxL seen in Rip7-BclxL/pIns-MYC-ERTAM (RM) double transgenic mice indicates that MYC-induced apoptosis is related to the Bax/Bak-mediated intrinsic mitochondrial pathway [27, 66]. Activation of this intrinsic apoptotic pathway was evident at the transcriptional level in β-cells by continued 2-fold increased expression of Bax and the somatic Cytochrome c gene (Cycs) from 16 hours onwards after MYC activation, and up-regulation of the mitochondrial respiratory gene for Endonuclease G, Endog, after only 4 hours of MYC activation. Both Bax[67, 68] and Cycs[42, 69] have been previously shown to be putative direct MYC targets due to the presence of non-canonical E-box MYC-Max binding sites, and association of MYC with the Bax promoter has been previously demonstrated through ChIP [68].
Gene expression profiling of pancreatic β-cells identified a strong and rapid induction of the DNA-damage response pathway (Table 2). A large increase in expression was detected after 8 hours of MYC activation for Rad51 and H2afx, previously identified MYC targets whose protein products are involved in homologous recombination and repair of DNA [70, 71]. Also, significant up-regulation of Hus1 and Rad1 - whose products form the 9-1-1 DNA-damage sensing machinery with Rad9 - indicated that oncogenic stress through deregulation of MYC resulted in the induction of DNA double strand breaks. The gene for the DNA-damage mediator Atr was also found to be up-regulated by 2-fold throughout the time course from 4 hours, and the associated checkpoint kinases Chk1 and Chk2 were up-regulated 2-fold from 8 hours (Table 2 and Figure 3C). The gene for the double-strand break-related DNA-damage mediator Atm showed 2-fold down-regulation at 8 hours, although it has been shown that Atm plays a significant role in MYC-induced apoptosis in lymphomagenesis in mice [72]. The genes Cdkn2a (p16/p19Arf) [2, 62] and Atr[73] are previously categorized MYC target genes. The checkpoint kinase genes Chk1 and Chk2 have not been previously classified as MYC target genes, indicating that the observed changes in expression may occur downstream of Atr. Interestingly, as mentioned previously, expression of Cdkn2c (p18Ink4c), which inhibits Cdk4/6, was significantly up-regulated (6-fold) in β-cells at 16 hrs. p18Ink4c interacts directly with Atm/Atr leading to an increase in p53 protein, and promotion of growth arrest and/or cell death, suggesting a link between p18 Ink4c and the DNA-damage response pathways [74]. Comparatively few DNA-damage related genes showed significant changes in expression for SBK, although we cannot exclude the possibility that DNA-damage proteins may be regulated by MYC through non-transcriptional means (e.g. protein phosphorylation).
Of particular interest was the survival factor gene Igf1, whose product has been shown to inhibit MYC-induced apoptosis in vitro by blocking Cytochrome c release from the mitochondria through the Akt1 tumour suppressor pathway [75]. Igf1 was up-regulated throughout much of the time-course for SBK, and this was confirmed using qRT-PCR (Figure 3C). Consistent with this finding, Akt1 was up-regulated 3-fold at 8 hours and 2-fold at 32 hours. A similar pattern was observed for Akt2, a second member of the Akt protein kinase family. This strongly supports the view that activation of the Igf-Akt pathway may contribute to suppression of MYC-related apoptosis. Interestingly, Igf1, Akt1 and Akt2 were up-regulated at later time points in β-cells. It is therefore possible that the Igf-Akt pathway is activated in both tissues, but that the β-cells may be able to bypass these signals.
The pro-apoptotic Bcl2 family member Pmaip1 (Noxa) and the gene encoding inhibitor of growth protein p47Ing3 (Ing3) both showed a decrease in expression of 2-fold at 8 hours in SBK, but exhibited no change in β-cells (Table 2). Over-expression of p47Ing3 results in cell cycle arrest and apoptosis in cancer cell lines [76], and reduced expression and loss of heterozygosity of the p47Ing3 locus have been found to be associated with human head and neck squamous carcinomas [77]. Also seen were several members of the TNF superfamily of apoptosis-inducing receptors. Tnfrsf12a, which has been found to be involved in inducing both apoptosis and angiogenesis [78, 79], showed an increase in expression in the skin of 3-fold at 8 hours. Tnfrsf4, whose product has been implicated in promoting survival through induction of Bcl2 and BclXL expression in CD4 T cells [80], similarly showed an increase in expression of 2-fold at 8 hours.
A pro-angiogenic response in suprabasal epidermis
The placental growth factor gene, Pgf¸ showed a marked increase from 8 hours throughout the time course for the skin (Table 3). In contrast, a 2-fold down-regulation was detected for the pancreas throughout much of the time-course. Pgf is a member of the vascular endothelial growth factor (VEGF) family, and has been shown to result in increased numbers, branching and size of dermal blood vessels following over-expression in basal keratinocytes of adult mice [81]. This may indicate a role in the development of neovasculature seen following MYC activation in the SBK [28]. Vegfa, a further member of the VEGF family important in angiogenesis, was down-regulated in the pancreas but showed up-regulation in the skin at 8 hours. In contrast, Vegfc, important in the growth of lymphatic vessels, showed down-regulation in the skin at early time points, but was up-regulated almost 4-fold in the pancreas at the later 32 hour time-point. Vegfb showed a 2-fold increase from 16 hours in pancreas compared to no change in the skin. These results indicate a transcriptional response upon MYC activation for genes relating to neovascular growth.
Activation of MYC leads to loss of differentiation
Activation of MYC is often associated with loss of differentiation of cells and has been found to block terminal differentiation in a variety of cell types [11, 82, 83]. Activation of MYC in the pancreatic β-cells resulted in down-regulation of Ins1 and Ins2 by > 5-fold at 4 hours, indicating acute loss of Insulin production within a short time period following MYC-deregulation. However, the expression levels of both subsequently increased dramatically showing almost 10-fold up-regulation from 16 hours (Table 3). Since the islet area used for RNA extraction was roughly identical for each sample, this indicates acute increase in the levels of Insulin production within the β-cells in response to continuous MYC activation, and not in response to an increase in β-cell mass. Although perhaps a paradox at first glance, this response may be the result of a positive feedback loop due to increased Insulin release into the bloodstream immediately following MYC activation as we have previously shown [23]. Observation of transcript levels of both mRNAs at a later time point (72 hours; data not shown) indicated that this period of high Insulin production is limited, as gene expression subsequently returned to lower levels indicative of loss of β-cell differentiation. This indicates a short window within the first two days where a balance is struck between an increased rate of Insulin production and the simultaneous loss of cells due to MYC-driven apoptosis.
Members of the homeodomain transcription factor family, Pdx1, Pax4, Hb9, Nkx2.2 and Nkx6.1, are essential in pancreatic development [84]. Probe-sets for the pancreatic and duodenal homeobox gene Pdx1 (or Ipf1), whose product activates transcription of the Insulin gene as well as a number of genes involved in glucose-sensing [85], showed a significant loss (> 3-fold) in expression at 8 hours following MYC activation (as well as potentially significant loss in expression < 2-fold at all other time points), which correlated with the early reduction seen in Insulin production (Table 3). The transcription factor gene Nkx6.1, whose product is essential for β-cell differentiation [86], also showed significant down-regulation in the early stages of MYC activation, although expression of this gene was shown to increase during later stages. Further Pdx1-regulated genes Slc2a2 (Glut2; previously classed as a putative MYC target gene) and Gck (Glucokinase), both part of the glucose-sensing machinery and involved in membrane transport and phosphorylation of glucose respectively, also followed similar expression profiles (Table 3). Gu et al. identified a group of 217 mature islet-specific genes [87], 63 of which showed a significant response following MYC activation in our analysis (Additional file 1, Table S6). The majority of these showed down-regulation at early time points. These data indicate a loss in β-cell differentiation and carbohydrate metabolism function following activation of MYC.
Activation of MYC in the SBK resulted in significant changes in expression of many genes relating to differentiation (Additional file 1, Table S5). In particular, it was clear that the primary result of MYC activation on these genes was down-regulation, with 199 differentiation-related genes showing a loss of expression compared to only 112 showing up-regulation. In addition to these general differentiation markers, activation of MYC led to down-regulation of several key keratinocyte differentiation genes (Table 3). Most notable was a significant 3-fold decrease in expression for the Involucrin gene, Ivl, after only 4 hours that was maintained throughout much of the time course. Involucrin is a key factor in the progression of differentiation of keratinocytes which works together with its substrate transglutaminase to cross-link with membrane proteins and provide support to the cell [88].
Cornifin (Sprr1b), a precursor to the epidermal cornified envelope, is a further keratinocyte differentiation markers that has been shown to affect the number of distinct layers of differentiated keratinocytes [89, 90]. As with Ivl, Sprr1b showed consistently marked down-regulation throughout the time course (Table 3). Similarly, Cystatin A (Csta), a cysteine protease inhibitor that is found expressed in keratinocytes as the precursor of the cornified cell envelope [91, 92], showed > 2-fold down-regulation throughout much of the time course. Up-regulation of α and β integrin genes such as Itga7, Itga9, Itgb2, Itgb3 and Itgb6, particularly at later time points, suggests altered adhesion of SBK with surrounding cells and the extracellular matrix following MYC activation (Table 3). Also, expression changes were detected for several Keratin genes, including up-regulation of the suprabasal-specific Krt1 and the basal-specific Krt14 at 8 hours (Table 3), which encode fibrous structural proteins in keratinocytes.
Previous findings from the Watt group in which MYC is targeted to basal keratinocytes has, in contrast, shown that activation of MYC promotes an increase in the number of proliferating keratinocytes concomitant with promotion of terminal differentiation of epidermal stem cells [89]. In the microarray experiment of Frye et al.[90], gene expression signatures were compared between whole skin sections from 4OHT-treated K14-MYC-ERTAM mice and 4OHT-treated WT mice to identify cellular networks involved in the promotion of terminal differentiation of epidermal stem cells at the expense of hair lineages [90]. Activation of MYC for 4 days was sufficient to cause hyperproliferation of the interfollicular epidermis, with increased expression of genes relating to both proliferation (e.g. Ki67, Krt17, Krt6, Cdc2, Cdc20) and interfollicular epidermis differentiation (Filaggrin and Cornifin/Sprp1). In contrast, expression of genes relating to early G1/S-phase cell cycle progression are more prominent when MYC is activated in SBK, whilst keratinocyte-specific differentiation genes such as Sprp1 generally show down-regulation. This suggests that activation of MYC suppresses differentiation in keratinocytes already undergoing terminal differentiation (i.e. SBK) to allow proliferation.
In a more recent study, low, intermediate, or high levels of MYC activity were induced in basal keratinocytes, and showed that MYC drives proliferation at all levels, but at high levels MYC promotes keratinocyte differentiation [93]. Promotion of differentiation may therefore act as a fail-safe mechanism against neoplastic conversion of epidermal stem cells.
The transcriptome fingerprint of MYC activation is varied in distinct tissues: pancreatic islets and skin
Clustering of genes whose expression changed significantly due to the joint effects of MYC activation and the tissue type identified genes whose expression profiles were correlated across the time courses for the two tissues, indicating possible co-regulation and functional similarity. Genes relating to DNA replication, DNA-damage checkpoint and cell cycle showed tight co-regulation, and showed significantly increased expression in the pancreatic β-cells. The expression profiles of mini-chromosome maintenance (MCM) deficient genes Mcm2, Mcm5, Mcm6 and Mcm7, whose products make up part of the MCM complex involved in DNA unwinding [94], the Cdt1 gene, whose product is involved in association of the MCM complex with chromatin, and various helicase related genes were found to be closely related, indicated co-expression of genes relating primarily to DNA replication. Close correlation with these genes was also seen in the pancreas for DNA-damage checkpoint related genes Atr and Chk1. These genes showed consistently high levels of expression in the β-cells, indicating a key role for DNA-damage response and repair in MYC-induced apoptosis. Conversely, no significant change was detected for these genes in the SBK.
In SBK, close correlation was detected for genes involved in proteolysis, particularly members of the Kallikrein family of serine proteases. Expression of Kallikrein genes (Klk1, Klk9, Klk21, Klk24 and Klk27) was found to increase significantly within 8 hours following activation of MYC, and continued to increase dramatically throughout the time course (Table 3 and Figure 1C). Klk1 and Klk9 have previously been found to be expressed throughout the epidermis of normal human skin [95], and play a role in degradation of the extracellular matrix (ECM) and loss of squamous cells during differentiation. Deregulated expression of Kallikreins has also been implicated in many cancer types [96–101]. The mouse Kallikreins Klk21, Klk24 and Klk27 have also been shown to be functionally active within the testes, both in degradation of the extra-cellular matrix and initiation of survival through degradation of Igf1bp3 [102–104]. Kallikrein proteins have been associated with angiogenesis. Growth of new vasculature is facilitated by regulating the tissue micro-environment, particularly through degradation of the extra-cellular matrix often via activation of matrix metalloproteinases (MMPs). This indicates a functional relationship between Kallikreins and the vascularisation-related gene, placental growth factor Pgf (a member of the vascular endothelial growth factor family of angiogenesis-related genes) and the Thrombospondin 2 gene Thbs2 (an anti-angiogenic factor which has been linked to extra-cellular matrix remodelling through modulation of matrix metalloproteinases [105]) which showed close correlation with these genes. Given the prominent angiogenic phenotype that develops in these pre-cancerous skin lesions following MYC activation [33], it is reasonable to speculate here that Kallikreins and Pgf may be involved.
Conclusions
Deregulation or over-expression of the MYC onco-protein is a frequent feature of human cancers, which attests to the pleiotropic role that ectopic MYC plays in cellular function. However, oncogenic MYC can also trigger activation of intrinsic tumour suppressor programs such as p19Arf/p53, which serve to limit propagation of such harmful cells by inducing growth arrest or apoptosis. While much is known about the mechanisms of MYC functions, the pathways responsible for deciding the ultimate fate of the cell between 'life' (uncontrolled proliferation) and 'death' (i.e. apoptosis) in vivo are not yet clear. The decision for a cell to become apoptotic depends on the complex interactions of many pro- and anti-apoptotic factors. Different tissues may exhibit varying levels of these factors ultimately determining the fate of a cell. However, tissue-specific environmental characteristics can also affect the interaction between these factors, having a decisive effect on cell fate. The MYC-ERTAM transgenic mouse model allows controlled over-expression of MYC in distinct adult tissues, SBK (epidermis) and pancreatic β-cells, enabling tracking of early changes downstream of aberrant MYC activity. In this study, high-throughput transcriptional profiling was used to identify transcriptional events that may provide clues to explain the disparity in the phenotypic response to MYC activation in SBK (proliferation and survival) and pancreatic β-cells (proliferation and apoptosis).
Whilst expression of genes relating to multiple cellular functions (eg. metabolic, transcriptional, transportational, ribosome biogenesis) is common to both tissues, expression of genes involved in DNA-damage and replication is enriched to β-cells. Consistent with early increased expression of Ki67 in β-cells [27], key G1/S-phase genes (e.g. those encoding Cyclin D2, Cyclin E and Cdk4) showed early changes in expression, whilst G2/M-phase genes (e.g. those encoding Cyclin A and Cyclin B) showed changes at later time points. Down-regulation of the cyclin-dependent kinase inhibitor (CDKI) Cdkn1b (p27Kip1) gene in β-cells was evident as previously shown in other cell types [48]. The CDKI Cdkn2c (p18Ink4c), which inhibits Cdk4 and Cdk6, was also down-regulated, consistent with G1/S phase transition. The short time period over which these changes were seen supports the idea that MYC-induced cell cycle progression occurs through direct activation of key cell cycle genes such as Ccnd1, Ccnd2 and Ccne2.
In SBK, although there were changes in cell cycle related genes, the response was much less prominent in comparison with β-cells. Gene expression changes included up-regulation of Krt6a, Pcna, Ccnd3, Cdk4, and the key G1/S phase cyclin gene Ccnd2, accompanied by significant down-regulation of the CDKI gene Cdkn1b (p27Kip1) throughout the time course. However, further cell cycle related genes (e.g. Ccna1, Ccne2 and Cdk2) did not appear to be measurably affected by MYC activation in SBKs. It is possible that SBK may at some point struggle to progress through the G2/M phase, which may be indicated by down-regulation of Ccnb1 and Cdc2a, whose products are essential in later cell cycle stages. This contrasts with pancreatic β-cells, in which we found both Ccnb1 and Cdc2a significantly up-regulated. The less pronounced cell cycle response in skin may be due to the relatively low proportion of keratinocytes that are responsive to Myc-induced proliferation at these early time-points. It has previously been shown that there is only a narrow window when very early suprabasal cells that have migrated out of the basal layer, are capable of responding to Myc-induced cell cycle entry [28, 105]. The more differentiated keratinocytes of the granular layer are refractory to the proliferative influence of MYC.
Gene expression profiling of the pancreatic β-cells identified the DNA-damage checkpoint pathway as a likely route by which MYC mediates apoptosis in this system, leading to downstream activation of p53 and Bax-mediated release of Cytochrome c from the mitochondria. In addition, close correlation was seen in the pancreas for DNA-damage checkpoint related genes Atr and Chk1, and members of the MCM complex, Mcm2, Mcm5 and Mcm7. The change in expression for these genes following MYC activation was consistently high in the β-cells, suggesting a key role for DNA-damage response and repair in MYC-induced apoptosis. Conversely, no significant change was detected for these genes in the SBK. Recent evidence strongly suggests that deregulated MYC induces rapid accumulation of DNA-damage, which is the primary cause of activation of the Atm/Atr-dependant checkpoint [106, 107]. The study of Dominguez-Sola et al. (2007), in particular, suggests that the pleiotropic role of MYC is due not only to transcriptional regulation of downstream genes, but also due to direct interactions with the DNA. This study also showed that over-expression of MYC results in DNA-damage and checkpoint activation. Consequently, the activation of DNA-damage response (DDR) pathways results in ultimate destruction of the offending cell.
Whilst direct control of these genes by MYC is not discernible from these data, it is clear that MYC deregulation induces a transcriptional response representative of cells undergoing DNA repair, which is a likely explanation for activation of the intrinsic apoptotic pathway. These results fit with the hypothesis that deregulated MYC leads to oncogenic stress and DNA-damage, although whether this is direct or indirect remains to be seen.
With regard to events downstream of the DDR, we found an increase in expression of genes associated with activation of mitochondrial outer membrane permeabilisation in pancreatic β-cells. These included the p53-target Bax, and the pro-apoptotic mitochondrial factors Cycs and Endog, which may indicate replenishment of proteins lost from the mitochondria during apoptotic signalling. In addition, increased expression of the tumour suppressor Cdkn2a (p19Arf) was also detected, which may have been involved in stabilization of the p53 tumour suppressor by inhibition of Mdm2, or in promoting cell cycle arrest prior to apoptosis together with up-regulation of the p53-target gene Cdkn1a (p21Cip1). In contrast, SBK showed no change in these genes, but instead a decrease in expression of the pro-apoptotic Bcl2 family member Pmaip1 (Noxa) and the inhibitor of growth gene Ing3 (p47Ing3). Previous work has linked over-expression of p47Ing3 with apoptosis [76], and reduced expression with human head and neck squamous carcinomas [77]. SBK may be protected from apoptosis in vivo by the Igf1-Akt survival pathway. Of particular interest was the early induction of Igf1, Akt1 and Akt2 in the SBK, and the tight correlation seen between the three. Expression of these three genes is also seen at later time points in the pancreas, indicating that the Igf1-Akt pathway may be activated in both tissues but is somehow bypassed in the β-cells.
One key difference between the two systems appears to be the presence of members of the Kallikrein serine protease family, which were dramatically up-regulated throughout the time-course for SBK. Kallikrein proteins have been linked to many forms of cancer, and of particular note is their role in the Igf1-Akt survival pathway. Klk1, Klk21, Klk24 and Klk27 have been linked to Igf1-regulated tumour survival through degradation of the Igf binding protein Igfbp3 in humans. This prevents Igfbp3 from antagonizing Igf1-Igf1r interactions, allowing Igf1 to bind to its receptor and initiate survival via the Akt pathway [102–104, 108]. Interestingly, the highest expression in a similar study from Frye et al. using a basal keratinocyte model for MYC activation [90] was for the brain and skin protease gene Bssp1, also known as Kallikrein 6 (Klk6). This gene remained low in WT treated mice, but was significantly increased following MYC expression (129-fold up-regulated). In both models, MYC activation drives vastly increased Kallikrein expression, so it is possible that these proteins play similar roles in cell survival in both systems. Increased expression of Kallikrein genes in SBK following MYC activation may therefore create an environment that favours survival over apoptosis.
In addition to the increased cell proliferation in both tissues, our data indicate a loss of differentiation in both pancreatic β-cells and SBK in response to activation of MYC. In pancreatic β-cells, we found down-regulation of genes that are essential in pancreatic development, such as Pdx1 and Nkx6.1, as well as genes involved in glucose-sensing such as Slc2a2 (Glut2) and Gck, both putative MYC-targets. In SBK, many significant changes were detected for genes relating to keratinocyte differentiation that generally pointed to a loss or delay in differentiation - an observation that was previously noted in this mouse model [28]. Interestingly, activation of MYC in the basal layer of the epidermis actually promotes keratinocyte differentiation [93], a mechanism which is thought to prevent epidermal stem cells from becoming cancerous. One possible explanation for the contrasting behaviour in suprabasal epidermis, is that SBKs are post-mitotic and have already entered a terminal differentiation process. Subsequent activation of MYC in early SBK may promote loss of differentiation to enable cell cycle entry. Since MYC-activated SBKs are already migrating upwards towards the skin surface, they are less likely to pose a cancer risk to the host given that they will ultimately be sloughed off as previously shown [28].
We have previously shown that MYC activation in SBK results in a prominent angiogenic phenotype [28]. From our microarray data, potential candidates that may promote such a response include Pgf, a member of the VEGF family, which was highly up-regulated in skin but in fact down-regulated in pancreas. We also found Vegfa up-regulated in skin but not in pancreas. Interestingly Vegfc, which is important in growth of lymphatic vessels, was down-regulated in skin but up-regulated in pancreas. However, given that prominent angiogenesis is not detected in the skin until 3-4 days following MYC activation [28], it is possible that the short time course considered here is too early to identify a transcriptional response in all relevant genes relating to vascularisation. Kallikrein proteins have also been implicated in facilitating angiogenesis via degradation of the cellular matrix [109] and our data showed co-regulation of Pgf and Kallikrein genes. These data suggest that the local tissue microenvironment in SBK that promote angiogenic growth may be linked to survival pathways that protect the cells against apoptosis.
In summary, activation of MYC results in cell growth, loss of differentiation and cell cycle entry in both β-cells and SBK in vivo. Apoptosis, which is confined to β-cells, involves a combination of a DNA-damage response and downstream activation of pro-apoptotic signalling pathways, including Cdc2a and p19Arf/p53, and downstream targets. Conversely, avoidance of apoptosis in SBK may result primarily from the activation of key anti-apoptotic signalling pathways, particularly those involved in the Igf1-Akt pathway, and induction of an angiogenic response (Pgf, Vegfa and Klks). A contributory role for intrinsic resistance to the induction of DNA-damage and the p19Arf tumour suppressor pathway by MYC in SBK is also possible. A possible mechanism whereby tissue-specific environmental factors may influence cell fate following MYC deregulation has also been proposed, hypothesising that the decision to live or die may relate to the local tissue-specific microenvironment. However, this remains speculation as the approach taken here gives an insight into only one aspect of the changes occurring within the cell in response to MYC deregulation. Much remains to be learnt from analysis of protein-level changes, post-translational modifications, or epigenetic modifications of DNA. This study has identified several new lines of investigation for future analysis into the dual roles of MYC in apoptosis and survival. It is hoped that such studies will prove fruitful and provide further insight into the complex role of this enigmatic protein.
Methods
Tissue Sample Preparation
Transgenic mice expressing switchable pIns-MYC-ERTAM in pancreatic β-cells or inv-MYC-ERTAM in SBK have been previously characterized and described [27, 28]. 8-12 week old male mice were treated with 4OHT or vehicle for 4, 8, 16 or 32 hours. Triplicate samples were collected for each time point for each of the four conditions; pIns-MYC-ERTAM 4OHT treated/MYC-ERTAM active ("Panc T"), pIns-MYC-ERTAM vehicle treated/MYC inactive ("Panc U"), Inv-MYC-ERTAM 4OHT treated/MYC active ("Skin T"), Inv-MYC-ERTAM vehicle treated/MYC inactive ("Skin U"). All mice were housed and treated in accordance with protocols and regulations sanctioned by the Home Office under the Animals Act of 1986.
RNA Isolation and Microarray Hybridization
A modified LCM protocol was devised to preserve RNA integrity. Fresh frozen pancreas sections were cut to a thickness of 15 μm, bound to a MMI MembraneSlide (Molecular Machines and Industries, Rockledge, FL) and fixed in ice-cold 100% ethanol for 2 mins. Sections were stained briefly (10 secs) with a 1% Toluidine Blue dye in 100%. Stained sections were dehydrated in 2 changes of 100% ethanol and 2 changes of xylene for 1 minute each, airdried for 2 minutes and finally left in a vacuum dessicator for 5 minutes before transportation to the laser capture platform. The SL μCut LCM system (Molecular Machines and Industries, Rockledge, FL) was used to isolate islets of Langerhans from surrounding exocrine tissue. RNA was collected using the RNA Microkit protocol (Qiagen, Valencia, CA). The laser capture procedure was repeated on freshly cut pancreas sections to collect a total area of islet cells equal to roughly 1.5 × 106 μm2 for each sample. Due to the thinness of murine suprabasal epidermis, isolation of sufficient good quality RNA for microarray hybridization from LCM of SBK proved difficult, a problem also noted by Agar et al. [110]. RNA was instead collected from 5 fresh frozen skin sections (20 μm) collected across several levels of the tissue. A modified version of the Affymetrix GeneChip 2-cycle target labelling in vitro transcription (IVT) protocol was used, incorporating double volumes of polyA controls and reagents in the first round cRNA synthesis stage to increase the yield. 10 μg double-amplified biotin-labelled cRNA were hybridized to Affymetrix MOE430 2.0 GeneChips (Affymetrix, Santa Clara, CA) as described in the Affymetrix GeneChip Expression Analysis Technical Manual.
Data Analysis
Data were normalized across all samples at the probe-level using GC-RMA [111]. Analysis of gene expression data was performed using the Bioconductor microarray analysis packages in R [112] and GeneSpring GX 7.3.1 (Agilent, Santa Clara, CA). 4OHT-treated samples were normalized to their respective vehicle-treated counterparts to ensure that normalized values corresponded to the fold-change in gene expression due to activation of MYC-ERTAM. Removal of control and non-responsive probes identified 12,349 probe sets. The custom R package Envisage was used to identify probe-sets showing significant differential expression after MYC-ERTAM activation, across time, and between pancreas and skin tissue. p-values were calculated for each model term and their interactions, and were corrected for multiple testing [113]. Only the highest-order interaction p-value was considered for genes where the selected model contains multiple terms relating to the MYC-ERTAM activation state. Contrast p-values were calculated for each condition by applying an unpaired t-test comparing 4OHT-treated samples with vehicle-treated samples within each of the 8 groups (4 hrs, 8 hrs, 16 hrs and 32 hrs for skin or pancreas). Significant early-changing (≥ 2-fold within 8 hours) probe sets were compared with known gene-ontologies using the DAVID functional annotation tool [29]. Quality threshold (QT) partitional clustering [114] was used to identify genes showing similar expression profiles, using the Pearson cross-correlation coefficient with a minimum correlation of 0.9 and a minimum cluster size of 14.
Quantitative Real-Time reverse transcriptase PCR
TaqMan qRT-PCR was performed on original total RNA samples for genes of interest. 20 ng total RNA was reverse transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). cDNA transcripts were pre-amplified prior to the qRT-PCR reaction in a multiplexed reaction using TaqMan preAmp mastermix (Applied Biosystems, Foster City, CA), with pooled TaqMan qRT-PCR assays (Applied Biosystems, Foster City, CA) at a concentration of 0.2X in 1X TE buffer. qRT-PCR was performed using an ABI Prism 7000 scanner (Applied Biosystems, Foster City, CA), with an 18s rRNA endogenous control probe (Applied Biosystems, Foster City, CA). qRT-PCR was performed for skin and pancreas 4OHT- and vehicle-treated RNA samples for early time-points 4 hrs and 8 hrs, and for the later 32 hrs time-point. As with microarray analysis, quantitative measures of gene expression upon MYC-activation were calculated by comparing vehicle- and 4OHT-treated samples directly for each condition.
Immunohistochemical Staining
Frozen sections were cut to 10 μm and fixed with 4% paraformaldehyde (PFA) at room temperature (RT) for 10 mins, washed in PBS for 5 mins, and incubated at RT in a humidifying temperature for 30 mins in 10% bovine serum albumin (BSA). Pancreas sections were double stained for Ki67 and insulin, or caspase 3 and insulin. Sections were incubated at 4°C overnight in primary antibodies diluted in 1% BSA: Insulin, 1:100 (guinea pig; DAKO, Denmark); Ki67, 1:200, (rabbit; Novocastra, UK); Caspase 3, 1:200 (rabbit; Cell Signalling Technology Inc., MA). Sections were washed twice in phosphate-buffered saline (PBS) with 0.1% tween (PBSt) for 5 mins each and incubated for 30 mins at RT in a humidifying chamber with secondary antibodies FITC (Vektor Co., Germany) or ALEXA633 (Invitrogen, Carlsbad, CA) diluted in 1% BSA (1:200). Skin tissue sections were sequentially stained for Keratin 1 and Ki67, or Keratin 1 and Caspase 3. Sections were incubated for 1 hour in Ki67 or Caspase 3 primary antibodies diluted in 1% BSA (1:200). Sections were washed twice with PBSt for 5 mins each and incubated for 30 mins at RT in a humidifying chamber with FITC anti-rabbit secondary antibodies diluted in 1% BSA (1:200). Finally, samples were washed in two changes of PBSt for 5 mins each. This cycle was repeated using Keratin 1 primary antibodies (rabbit; BabCo, CA) diluted in 1% BSA (1:100) and ALEXA633 anti-rabbit secondary antibodies diluted in 1% BSA (1:200). Slides were mounted in Vectashield (Vector Labs, Burlingame, CA) mounting medium containing 4',6-diamidino-2-phenylindole (DAPI) and viewed using a Leica Sp2 confocal microscope (Leica, Wetzlar, Germany).
Abbreviations
- 4OHT:
-
4-Hydroxytamoxifen
- 2a-cRNA:
-
Double-amplified biotin-labelled complementary RNA
- ANOVA:
-
Analysis of Variance
- BSA:
-
Bovine Serum Albumin
- CDK:
-
Cyclin-Dependent Kinase
- CDKI:
-
Cyclin-Dependent Kinase Inhibitor
- ChIP:
-
Chromatin Immunoprecipitation
- cRNA:
-
Complementary RNA
- DAPI:
-
4',6-diamidino-2-phenylindole
- DNA:
-
Deoxyribonucleic Acid
- DPX:
-
Distyrene/Plasticizer/Xylene
- ECM:
-
Extra Cellular Matrix
- FDR:
-
False Discovery Rate
- FWER:
-
Family Wise Error Rate
- GCOS:
-
Genechip Operating System
- GC-RMA:
-
Genechip Robust Multi-array Average
- GO:
-
Gene Ontology
- IVT:
-
In-Vitro Transcription
- LCM:
-
Laser Capture Microdissection
- MCM:
-
Minichromosome Maintenance
- MMP:
-
Matrix Metalloproteinase
- OCT:
-
Optimal Cutting Temperature
- PBS:
-
Phosphate Buffered Saline
- PFA:
-
Paraformaldehyde
- QC:
-
Quality Control
- qRT-PCR:
-
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction
- RIN:
-
RNA Integrity Number
- RM:
-
RIP7-BclXL/pIns-MYC-ERTAM
- RNA:
-
Ribonucleic Acide
- RT:
-
Room Temperature
- SAGE:
-
Serial Analysis of Gene Expression
- SBK:
-
Skin Basal Keratinocytes
- VEGF:
-
Vascular Endothelial Growth Factor
References
Amati B, Alevizopoulos K, Vlach J: Myc and the cell cycle. Front Biosci. 1998, 3: d250-268.
Dang CV: c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999, 19 (1): 1-11.
Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR: Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000, 97 (7): 3260-3265. 10.1073/pnas.97.7.3260.
de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW: Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001, 14 (1): 45-55. 10.1016/S1074-7613(01)00088-7.
Eisenman RN: Deconstructing myc. Genes Dev. 2001, 15 (16): 2023-2030. 10.1101/gad928101.
Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM: c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001, 414 (6865): 768-773. 10.1038/414768a.
Pelengaris S, Khan M, Evan G: c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002, 2 (10): 764-776. 10.1038/nrc904.
Pelengaris S, Khan M: The many faces of c-MYC. Arch Biochem Biophys. 2003, 416 (2): 129-136. 10.1016/S0003-9861(03)00294-7.
Persson H, Hennighausen L, Taub R, DeGrado W, Leder P: Antibodies to human c-myc oncogene product: evidence of an evolutionarily conserved protein induced during cell proliferation. Science. 1984, 225 (4663): 687-693. 10.1126/science.6431612.
Henriksson M, Luscher B: Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996, 68: 109-182.
Freytag SO: Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distinct predifferentiation state in G0/G1. Mol Cell Biol. 1988, 8 (4): 1614-1624.
Hoffman-Liebermann B, Liebermann DA: Interleukin-6- and leukemia inhibitory factor-induced terminal differentiation of myeloid leukemia cells is blocked at an intermediate stage by constitutive c-myc. Mol Cell Biol. 1991, 11 (5): 2375-2381.
Selvakumaran M, Liebermann D, Hoffman-Liebermann B: Myeloblastic leukemia cells conditionally blocked by myc-estrogen receptor chimeric transgenes for terminal differentiation coupled to growth arrest and apoptosis. Blood. 1993, 81 (9): 2257-2262.
Wagner AJ, Kokontis JM, Hay N: Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994, 8 (23): 2817-2830. 10.1101/gad.8.23.2817.
Tanaka H, Matsumura I, Ezoe S, Satoh Y, Sakamaki T, Albanese C, Machii T, Pestell RG, Kanakura Y: E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol Cell. 2002, 9 (5): 1017-1029. 10.1016/S1097-2765(02)00522-1.
Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM: c-Myc can induce DNA-damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002, 9 (5): 1031-1044. 10.1016/S1097-2765(02)00520-8.
Alarcon RM, Rupnow BA, Graeber TG, Knox SJ, Giaccia AJ: Modulation of c-Myc activity and apoptosis in vivo. Cancer Res. 1996, 56 (19): 4315-4319.
Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjo S, Murga M, et al: Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010, 12 (1): 54-59. 10.1038/ncb2004. sup pp 51-14
van Riggelen J, Felsher DW: Myc and a Cdk2 senescence switch. Nat Cell Biol. 2010, 12 (1): 7-9. 10.1038/ncb0110-7.
Soucek L, Evan GI: The ups and downs of Myc biology. Curr Opin Genet Dev. 2010, 20 (1): 91-95. 10.1016/j.gde.2009.11.001.
Harrington EA, Fanidi A, Evan GI: Oncogenes and cell death. Curr Opin Genet Dev. 1994, 4 (1): 120-129. 10.1016/0959-437X(94)90100-7.
Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV: An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003, 4 (10): R69-10.1186/gb-2003-4-10-r69.
Cheung L, Zervou S, Mattsson G, Abouna S, Zhou L, Ifandi V, Pelengaris S, Khan M: c-Myc directly induces both impaired insulin secretion and loss of β-cell mass, independently of hyperglycemia in vivo. Islets. 2010, 2 (1): 37-45. 10.4161/isl.2.1.10196.
Menssen A, Hermeking H: Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002, 99 (9): 6274-6279. 10.1073/pnas.082005599.
Lewis BC, Shim H, Li Q, Wu CS, Lee LA, Maity A, Dang CV: Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997, 17 (9): 4967-4978.
Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB: Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004, 4 (7): 562-568. 10.1038/nrc1393.
Pelengaris S, Khan M, Evan GI: Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002, 109 (3): 321-334. 10.1016/S0092-8674(02)00738-9.
Pelengaris S, Littlewood T, Khan M, Elia G, Evan G: Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999, 3 (5): 565-577. 10.1016/S1097-2765(00)80350-0.
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA: DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4 (5): P3-10.1186/gb-2003-4-5-p3.
van Riggelen J, Yetil A, Felsher DW: MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010, 10 (4): 301-309. 10.1038/nrc2819.
Finch A, Prescott J, Shchors K, Hunt A, Soucek L, Dansen TB, Swigart LB, Evan GI: Bcl-xL gain of function and p19 ARF loss of function cooperate oncogenically with Myc in vivo by distinct mechanisms. Cancer Cell. 2006, 10 (2): 113-120. 10.1016/j.ccr.2006.06.017.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005, 102 (43): 15545-15550. 10.1073/pnas.0506580102.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al: Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006, 439 (7074): 353-357. 10.1038/nature04296.
Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS: Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004, 36 (12): 1306-1311. 10.1038/ng1481.
Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al: The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001, 29 (2): 397-406. 10.1093/nar/29.2.397.
Yu Q, He M, Lee NH, Liu ET: Identification of Myc-mediated death response pathways by microarray analysis. J Biol Chem. 2002, 277 (15): 13059-13066. 10.1074/jbc.M111403200.
Eilers M: Control of cell proliferation by Myc family genes. Mol Cells. 1999, 9 (1): 1-6.
Flores I, Murphy DJ, Swigart LB, Knies U, Evan GI: Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene. 2004, 23 (35): 5923-5930. 10.1038/sj.onc.1207796.
Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM: Over-expression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci USA. 2000, 97 (19): 10544-10548.
Jansen-Durr P, Meichle A, Steiner P, Pagano M, Finke K, Botz J, Wessbecher J, Draetta G, Eilers M: Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci USA. 1993, 90 (8): 3685-3689. 10.1073/pnas.90.8.3685.
Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M: Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994, 14 (6): 4032-4043.
Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET: Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000, 60 (21): 5922-5928.
Yin XY, Grove L, Datta NS, Katula K, Long MW, Prochownik EV: Inverse regulation of cyclin B1 by c-Myc and p53 and induction of tetraploidy by cyclin B1 over-expression. Cancer Res. 2001, 61 (17): 6487-6493.
Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M: Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. Embo J. 1999, 18 (19): 5321-5333. 10.1093/emboj/18.19.5321.
Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H: Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1). Embo J. 1999, 18 (19): 5310-5320. 10.1093/emboj/18.19.5310.
Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B: Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001, 15 (16): 2042-2047. 10.1101/gad.907901.
Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al: Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000, 97 (5): 2229-2234. 10.1073/pnas.050586197.
Yang W, Shen J, Wu M, Arsura M, FitzGerald M, Suldan Z, Kim DW, Hofmann CS, Pianetti S, Romieu-Mourez R, et al: Repression of transcription of the p27(Kip1) cyclin-dependent kinase inhibitor gene by c-Myc. Oncogene. 2001, 20 (14): 1688-1702. 10.1038/sj.onc.1204245.
Th'ng JP, Wright PS, Hamaguchi J, Lee MG, Norbury CJ, Nurse P, Bradbury EM: The FT210 cell line is a mouse G2 phase mutant with a temperature-sensitive CDC2 gene product. Cell. 1990, 63 (2): 313-324. 10.1016/0092-8674(90)90164-A.
Itzhaki JE, Gilbert CS, Porter AC: Construction by gene targeting in human cells of a "conditional' CDC2 mutant that rereplicates its DNA. Nat Genet. 1997, 15 (3): 258-265.
Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Caceres JF, Dubus P, Malumbres M, Barbacid M: Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007, 448 (7155): 811-815. 10.1038/nature06046.
Kaldis P, Aleem E: Cell cycle sibling rivalry: Cdc2 vs. Cdk2. Cell Cycle. 2005, 4 (11): 1491-1494. 10.4161/cc.4.11.2124.
Castedo M, Perfettini JL, Roumier T, Kroemer G: Cyclin-dependent kinase-1: linking apoptosis to cell cycle and mitotic catastrophe. Cell Death Differ. 2002, 9 (12): 1287-1293. 10.1038/sj.cdd.4401130.
Serrano M, Hannon GJ, Beach D: A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993, 366 (6456): 704-707. 10.1038/366704a0.
Guan KL, Jenkins CW, Li Y, Nichols MA, Wu X, O'Keefe CL, Matera AG, Xiong Y: Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994, 8 (24): 2939-2952. 10.1101/gad.8.24.2939.
Goruppi S, Ruaro E, Schneider C: Gas6, the ligand of Axl tyrosine kinase receptor, has mitogenic and survival activities for serum starved NIH3T3 fibroblasts. Oncogene. 1996, 12 (3): 471-480.
Li R, Chen J, Hammonds G, Phillips H, Armanini M, Wood P, Bunge R, Godowski PJ, Sliwkowski MX, Mather JP: Identification of Gas6 as a growth factor for human Schwann cells. J Neurosci. 1996, 16 (6): 2012-2019.
Ganopolsky JG, Abid MR, Aird WC, Blostein MD: GAS6-induced signaling in human endothelial cells is mediated by FOXO1a. J Thromb Haemost. 2008, 6 (10): 1804-1811. 10.1111/j.1538-7836.2008.03114.x.
Shankar SL, O'Guin K, Cammer M, McMorris FA, Stitt TN, Basch RS, Varnum B, Shafit-Zagardo B: The growth arrest-specific gene product Gas6 promotes the survival of human oligodendrocytes via a phosphatidylinositol 3-kinase-dependent pathway. J Neurosci. 2003, 23 (10): 4208-4218.
Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ: Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002, 277 (40): 36921-36930. 10.1074/jbc.M201493200.
Jensen KB, Watt FM: Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci USA. 2006, 103 (32): 11958-11963. 10.1073/pnas.0601886103.
Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF: Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998, 12 (15): 2424-2433. 10.1101/gad.12.15.2424.
Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL: Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999, 13 (20): 2658-2669. 10.1101/gad.13.20.2658.
Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI: Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008, 14 (6): 447-457. 10.1016/j.ccr.2008.10.018.
Halgren RG, Fielden MR, Fong CJ, Zacharewski TR: Assessment of clone identity and sequence fidelity for 1189 IMAGE cDNA clones. Nucleic Acids Res. 2001, 29 (2): 582-588. 10.1093/nar/29.2.582.
Dansen TB, Whitfield J, Rostker F, Brown-Swigart L, Evan GI: Specific requirement for Bax, not Bak, in Myc-induced apoptosis and tumor suppression in vivo. J Biol Chem. 2006, 281 (16): 10890-10895. 10.1074/jbc.M513655200.
Mitchell KO, Ricci MS, Miyashita T, Dicker DT, Jin Z, Reed JC, El-Deiry WS: Bax is a transcriptional target and mediator of c-myc-induced apoptosis. Cancer Res. 2000, 60 (22): 6318-6325.
Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B: Genomic targets of the human c-Myc protein. Genes Dev. 2003, 17 (9): 1115-1129. 10.1101/gad.1067003.
Morrish F, Giedt C, Hockenbery D: c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003, 17 (2): 240-255. 10.1101/gad.1032503.
Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B: A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA. 2003, 100 (14): 8164-8169. 10.1073/pnas.1332764100.
Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, Farnham PJ, Huang TH, Penn LZ: Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol. 2003, 13 (10): 882-886. 10.1016/S0960-9822(03)00297-5.
Maclean KH, Kastan MB, Cleveland JL: Atm deficiency affects both apoptosis and proliferation to augment Myc-induced lymphomagenesis. Mol Cancer Res. 2007, 5 (7): 705-711. 10.1158/1541-7786.MCR-07-0058.
Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D: A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003, 31 (21): 6148-6156. 10.1093/nar/gkg794.
Park BJ, Kang JW, Lee SW, Choi SJ, Shin YK, Ahn YH, Choi YH, Choi D, Lee KS, Kim S: The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell. 2005, 120 (2): 209-221. 10.1016/j.cell.2004.11.054.
Lowe SW, Cepero E, Evan G: Intrinsic tumour suppression. Nature. 2004, 432 (7015): 307-315. 10.1038/nature03098.
Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, Yokota J, Harris CC: A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene. 2003, 22 (3): 343-350. 10.1038/sj.onc.1206115.
Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, Nagai N, Nishizaki K, Shimizu K: Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene. 2002, 21 (28): 4462-4470. 10.1038/sj.onc.1205540.
Wiley SR, Cassiano L, Lofton T, Davis-Smith T, Winkles JA, Lindner V, Liu H, Daniel TO, Smith CA, Fanslow WC: A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001, 15 (5): 837-846. 10.1016/S1074-7613(01)00232-1.
Wiley SR, Winkles JA: TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003, 14 (3-4): 241-249. 10.1016/S1359-6101(03)00019-4.
Rogers PR, Song J, Gramaglia I, Killeen N, Croft M: OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity. 2001, 15 (3): 445-455. 10.1016/S1074-7613(01)00191-1.
Odorisio T, Schietroma C, Zaccaria ML, Cianfarani F, Tiveron C, Tatangelo L, Failla CM, Zambruno G: Mice overexpressing placenta growth factor exhibit increased vascularization and vessel permeability. J Cell Sci. 2002, 115 (Pt 12): 2559-2567.
Coppola JA, Cole MD: Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986, 320 (6064): 760-763. 10.1038/320760a0.
Maruyama K, Schiavi SC, Huse W, Johnson GL, Ruley HE: myc and E1A oncogenes alter the responses of PC12 cells to nerve growth factor and block differentiation. Oncogene. 1987, 1 (4): 361-367.
Chakrabarti SK, Mirmira RG: Transcription factors direct the development and function of pancreatic beta cells. Trends Endocrinol Metab. 2003, 14 (2): 78-84. 10.1016/S1043-2760(02)00039-5.
Hui H, Perfetti R: Pancreas duodenum homeobox-1 regulates pancreas development during embryogenesis and islet cell function in adulthood. Eur J Endocrinol. 2002, 146 (2): 129-141. 10.1530/eje.0.1460129.
Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M: Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000, 127 (24): 5533-5540.
Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, Melton DA: Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development. 2004, 131 (1): 165-179. 10.1242/dev.00921.
Eckert RL, Green H: Structure and evolution of the human involucrin gene. Cell. 1986, 46 (4): 583-589. 10.1016/0092-8674(86)90884-6.
Arnold I, Watt FM: c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001, 11 (8): 558-568. 10.1016/S0960-9822(01)00154-3.
Frye M, Gardner C, Li ER, Arnold I, Watt FM: Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003, 130 (12): 2793-2808. 10.1242/dev.00462.
Jarvinen M, Rinne A, Hopsu-Havu VK: Human cystatins in normal and diseased tissues--a review. Acta Histochem. 1987, 82 (1): 5-18.
Hawley-Nelson P, Roop DR, Cheng CK, Krieg TM, Yuspa SH: Molecular cloning of mouse epidermal cystatin A and detection of regulated expression in differentiation and tumorigenesis. Mol Carcinog. 1988, 1 (3): 202-211. 10.1002/mc.2940010309.
Berta MA, Baker CM, Cottle DL, Watt FM: Dose and context dependent effects of Myc on epidermal stem cell proliferation and differentiation. EMBO Mol Med. 2010, 2 (1): 16-25. 10.1002/emmm.200900047.
Ishimi Y: A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997, 272 (39): 24508-24513. 10.1074/jbc.272.39.24508.
Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, Saijoh K: Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J Invest Dermatol. 2003, 121 (3): 542-549. 10.1046/j.1523-1747.2003.12363.x.
Borgono CA, Diamandis EP: The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004, 4 (11): 876-890. 10.1038/nrc1474.
Petraki CD, Gregorakis AK, Papanastasiou PA, Karavana VN, Luo LY, Diamandis EP: Immunohistochemical localization of human kallikreins 6, 10 and 13 in benign and malignant prostatic tissues. Prostate Cancer Prostatic Dis. 2003, 6 (3): 223-227. 10.1038/sj.pcan.4500674.
Sano A, Sangai T, Maeda H, Nakamura M, Hasebe T, Ochiai A: Kallikrein 11 expressed in human breast cancer cells releases insulin-like growth factor through degradation of IGFBP-3. Int J Oncol. 2007, 30 (6): 1493-1498.
Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, et al: Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003, 63 (24): 8614-8622.
Yousef GM, Yacoub GM, Polymeris ME, Popalis C, Soosaipillai A, Diamandis EP: Kallikrein gene down-regulation in breast cancer. Br J Cancer. 2004, 90 (1): 167-172. 10.1038/sj.bjc.6601451.
Yousef GM, Kyriakopoulou LG, Scorilas A, Fracchioli S, Ghiringhello B, Zarghooni M, Chang A, Diamandis M, Giardina G, Hartwick WJ, et al: Quantitative expression of the human kallikrein gene 9 (KLK9) in ovarian cancer: a new independent and favorable prognostic marker. Cancer Res. 2001, 61 (21): 7811-7818.
Matsui H, Moriyama A, Takahashi T: Cloning and characterization of mouse klk27, a novel tissue kallikrein expressed in testicular Leydig cells and exhibiting chymotrypsin-like specificity. Eur J Biochem. 2000, 267 (23): 6858-6865. 10.1046/j.1432-1033.2000.01786.x.
Matsui H, Takahashi T: Mouse testicular Leydig cells express Klk21, a tissue kallikrein that cleaves fibronectin and IGF-binding protein-3. Endocrinology. 2001, 142 (11): 4918-4929. 10.1210/en.142.11.4918.
Matsui H, Takano N, Moriyama A, Takahashi T: Single-chain tissue-type plasminogen activator is a substrate of mouse glandular kallikrein 24. Zoolog Sci. 2005, 22 (10): 1105-1111. 10.2108/zsj.22.1105.
Krady MM, Zeng J, Yu J, MacLauchlan S, Skokos EA, Tian W, Bornstein P, Sessa WC, Kyriakides TR: Thrombospondin-2 modulates extracellular matrix remodeling during physiological angiogenesis. Am J Pathol. 2008, 173 (3): 879-891. 10.2353/ajpath.2008.080128.
Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R: Non-transcriptional control of DNA replication by c-Myc. Nature. 2007, 448 (7152): 445-451. 10.1038/nature05953.
Pusapati RV, Rounbehler RJ, Hong S, Powers JT, Yan M, Kiguchi K, McArthur MJ, Wong PK, Johnson DG: ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci USA. 2006, 103 (5): 1446-1451. 10.1073/pnas.0507367103.
Rehault S, Monget P, Mazerbourg S, Tremblay R, Gutman N, Gauthier F, Moreau T: Insulin-like growth factor binding proteins (IGFBPs) as potential physiological substrates for human kallikreins hK2 and hK3. Eur J Biochem. 2001, 268 (10): 2960-2968. 10.1046/j.1432-1327.2001.02185.x.
Tschesche H, Michaelis J, Kohnert U, Fedrowitz J, Oberhoff R: Tissue kallikrein effectively activates latent matrix degrading metalloenzymes. Adv Exp Med Biol. 1989, 247A: 545-548.
Agar NS, Halliday GM, Barnetson RS, Jones AM: A novel technique for the examination of skin biopsies by laser capture microdissection. J Cutan Pathol. 2003, 30 (4): 265-270. 10.1046/j.0303-6987.2003.052.x.
Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F: A Model Based Background Adjustment for Oligonucleotide Expression Arrays. Johns Hopkins University Department of Biostatistics Working Papers. 2004
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al: Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004, 5 (10): R80-10.1186/gb-2004-5-10-r80.
Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995, Ser. B 57: 289-300.
Heyer LJ, Kruglyak S, Yooseph S: Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999, 9 (11): 1106-1115. 10.1101/gr.9.11.1106.
Acknowledgements
The authors wish to thank Giorgia Riboldi-Tunnicliffe of Affymetrix UK Ltd. for her assistance with sample preparation optimisation, Geoff Scopes of Affymetrix UK Ltd. for his assistance with array quality control procedures, and Jane Hadlington and Helen Bottomley of Applied Biosystems Ltd. for their assistance with qRT-PCR optimisation. Funding for this project was jointly provided by the EPSRC, BBSRC, AICR, Wellcome Trust, Eli Lilly & Co. Ltd., and Amylin Pharmaceuticals, Inc.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
At the time this work was carried out, EH was Senior Scientist in Bioinformatics (EMEA) for Agilent Technologies, the developers of the GeneSpring GX analysis suite. The remaining authors declare that they have no competing interests.
Authors' contributions
SR conducted the major part of this study, including designing the experiment, preparing samples, developing the Envisage tools, running all statistical analyses, performing qRT-PCR and immunohistochemical staining, and drafting the manuscript. LW performed all microarray hybridisations. HB assisted with experiment design, microarray hybridisation and manuscript preparation. HT and EH assisted with development of Envisage and other statistical analyses. SP and MK conceived of the study, coordinated the study design, and helped to finalise the manuscript. All authors read and approved the final manuscript.
Data Access
The microarray data described in this experiment are available from the ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MEXP-2952. Envisage code is available upon request.
Stella Pelengaris and Michael Khan contributed equally to this work.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Robson, S.C., Ward, L., Brown, H. et al. Deciphering c-MYC-regulated genes in two distinct tissues. BMC Genomics 12, 476 (2011). https://doi.org/10.1186/1471-2164-12-476
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-12-476