Abstract
Background
Repetitive sequences are the major components of the eukaryotic genomes. Association of these repeats with transcribing sequences and their regulation in buffalo Bubalus bubalis has remained largely unresolved.
Results
We cloned and sequenced RsaI repeat fragments pDp1, pDp2, pDp3, pDp4 of 1331, 651, 603 and 339 base pairs, respectively from the buffalo, Bubalus bubalis. Upon characterization, these fragments were found to represent retrotransposons and part of some functional genes. The resultant clones showed cross hybridization only with buffalo, cattle, goat and sheep genomic DNA. Real Time PCR, detected ~2 × 104 copies of pDp1, ~ 3000 copies of pDp2 and pDp3 and ~ 1000 of pDp4 in buffalo, cattle, goat and sheep genomes, respectively. RsaI repeats are transcriptionally active in somatic tissues and spermatozoa. Accordingly, pDp1 showed maximum expression in lung, pDp2 and pDp3 both in Kidney, and pDp4 in ovary. Fluorescence in situ hybridization showed repeats to be distributed all across the chromosomes.
Conclusions
The data suggest that RsaI repeats have been incorporated into the exonic regions of various transcribing genes, possibly contributing towards the architecture and evolution of the buffalo and related genomes. Prospects of our present work in the context of comparative and functional genomics are highlighted.
Similar content being viewed by others
Background
Different families of repetitive DNA contribute towards architectural organization of the mammalian genomes [1]. They represent both, tandemly arranged and interspersed sequences [2]. Based on their size and mode of propagation, Interspersed elements can be divided into two separate classes, the long terminal repeat (LTR) and non-LTR. The non LTR LINEs (long interspersed repeat elements) and SINEs (Short interspersed repeat elements) are widely distributed occupying a substantial fraction of the eukaryotic genomes. These elements replicate and proliferate themselves through a "copy and paste" mechanism called retrotransposition [3, 4]. In this process, transcription of their genomic copies is followed by an RNA intermediate resulting cDNAs reintegration at a new location in the genome [5]. Approximately, 100 LINE and SINE families have been reported to date in various eukaryotic genomes [6]. In mammals, LINE, L1 repeats are dominant retrotransposons type both in the common ancestor and in extant species [7]. In addition to L1, an element belonging to the retrotransposable element family of autonomous retrotransposons (RTE-1) has been reported in mammals [8]. Few mammals have active non LTR LINE other than L1 that contribute significantly to repeat composition.
Species specific retrotransposons have been widely used as a tool for Phylogenetic analysis and population studies [9, 10]. Many retrotransposons are inactive, found in the non-coding regions of the genome and are subjected only to the neutral evolution. Thus, rare new insertions have led to some form of advantageous or noteworthy phenotypic variations [11–13]. These and other such discoveries have resulted in a shift from earlier thought of them being "parasite" to functional elements cultivated in the genome for their beneficial attributes.
Retrotransposons have gained novel functions, providing alternative splice sites and/or polyadenylation signals or modifying gene expression [14–16]. These elements account for 46.5% of bovine genome [17]. The bovine genome is very different in its repeat composition compared to other mammalian genomes. It has unusual composition of LINE RTE type Bov B and its associated SINE elements which together account for 25% of the bovine genome [17]. The impact of the interspersed repeats on the genomes of human [18–20], dog [21, 22], cow [17, 23], mouse [24] and opossum [25, 26] has been studied. However fate of these interspersed elements and their association with mRNA transcriptomes in buffalo remains still unclear. Here, we report RsaI family repetitive DNA in the genome of water buffalo "Bubalus bubalis" and their copy number status. We also studied their expression in somatic tissues and spermatozoa. The repeat fraction pDp1, pDp2 and pDp3 were used for fluorescence in situ hybridization (FISH) with buffalo metaphase chromosomes. In addition, we isolated and sequenced ACOT11 ( Acyl-coenzyme A thioesterase 11) gene harboring part of pDp1 repeat.
Results
RsaI enzyme digestion uncovers four repeat fractions
Digestion of buffalo genomic DNA with RsaI enzyme, besides minor ones, showed four prominent bands ranging from 1331 base pairs, pDp1; 651, pDp2; 603, pDp3; to 339; pDp4 (Figure 1). Approximately, 15-20 recombinant clones subjected to restriction digestion and slot blot hybridization screening yielded ten positive clones for each repeat. Five-six clones from each fragment were then sequenced. The accession numbers of the recombinant clones are given in the Additional File 1. All the four major repeat elements were AT rich but sequence-wise, were different from one another (Additional File 2). No inter-clonal variations were detected in these sequences. However, random repeats were present in the nucleotide sequences (Table 1). Repeat Masker programme revealed presence of LTR LINE, SINE elements within the four fragments (Table 2). Blast search for each clone showed 69-98% homology with genomic DNA/contigs and 68-93% with transcribing genes in the database (Additional File 1) mostly in UTRs (Additional File 3).
Buffalo derived RsaI Open Reading Frame (ORFs) has amino acid similarity to LINE reverse transcriptase
Most striking feature of pDp1 sequence was the presence of 489 bp, +1 ORF (nucleotide position, 841-1329) (Figure 2B). BLASTP search with GenBank sequences using conceptual translation of this ORF (162aa) gave matches to putative reverse transcriptase's domain. This region corresponds to central position of the Reverse transcriptase ORF (Figures 2A and 3). Similarly, pDp2, 111 bp, +3 ORF (nucleotide position, 135-245, 65aa) and pDp4, 135 bp, +3 ORF (nucleotide position, 81-215, 44aa) showed homology to endonuclease reverse transcriptase (Figures 2B and 2C). These ORFs corresponded to central endonuclease reverse transcriptase of LINE1 ORF2 (Figure 3). pDp3 sequence,112 bp, +3 ORF (nucleotide position, 492-602, 37 aa) showed no similarity with endonuclease reverse transcriptase.
Amino acid alignment of buffalo derived RsaI sequences showing positions of pDp1, 2 and 4 based on ORF search. Panel (A) is a diagrammatic illustration showing 5'UTR, ORF1, Endonuclease, Reverse transcriptase and 3'UTS of LINE. Position of pDp1 (Blue), pDp2 and pDp4 (Red) within LINE are shown on top of the figure. The aligned sequences in the panel (B) shows region with homology to reverse transcriptase domain (Blue). Panel ( C) and (D), shows region with homology to endonuclease domain (Red).
PCR amplification of bovine genomic DNA using internal primers designed from the ORF of buffalo RsaI sequences. Schematic representation (A) shows PCR strategy used for amplification of ORF regions of RsaI sequences corresponding to 489, 141, 112 and 135 bp, respectively (a-d). Panel (B) shows PCR amplification of RsaI ORF regions and β-actin as control. The corresponding position of each PCR product is shown in Panel (A). Sequence IDs are indicated on left, amplicons size on right and species are mentioned on top of the lanes.
RsaI Repeat status among bovids
Independent cross hybridization of pDp1, pDp2, pDp3 and pDp4 with genomic DNA from different species (mentioned in methods section) under high stringent conditions showed signals only in bovids (Additional File 4). PCR conducted using region specific primers (Table 3) amplified bands in buffalo, cattle, goat and sheep genomic DNA (Figures 3A & 3B). Southern hybridization of two representative RsaI sequence pDp1, and pDp2 showed same number of bands within the genomic DNA of bovids (Additional File 5). Prominent bands within the smear reflect identical size of the fragments dispersed throughout the bovid genomes. ClustalW alignment of 489 bp pDp1derived fragment with cattle (AF060172), goat (AF404302) and sheep (AC148038) showed high level of sequence conservation among the bovids (Additional File 6). Phylogenetically, with respect to all the four sequences, cattle and buffalo were found to be closer to each other (Additional File 7), constituting the same monophyletic group.
Differential expression of RsaI fragments in somatic tissue and spermatozoa of buffalo
RT-PCR analysis using internal primers of pDp1, pDp2, pDp3 and pDp4 (see Table 3) and cDNA from somatic tissues and spermatozoa of buffalo showed amplification of band across tissues and sperm confirming their transcriptional potentials (Additional File 8). Quantitative Real Time PCR analysis of these sequences showed differential expression of RsaI related transcripts across somatic tissues and spermatozoa (Figure 4). pDp1 showed highest expression in lung, pDp2 and pDp3 in Kidney and pDp4 in ovary. Summary of the relative expression (in folds) derived from 2-ΔΔCtvalues obtained for various transcripts based on Real Time PCR are given in Table 4.
Multiple copies of RsaI fragments in bovids
The pair of internal primers deduced from pDp1-4 used for expression study was also employed for copy number analysis of these fragments in cattle buffalo, goat and sheep using Real Time PCR. Standard curve slope value was between 3.2-3.5. Single melting peak on dissociate curve confirmed primer specificity. pDp1 showed ~2 × 104 copies, pDp2 and pDp3, ~ 3000 copies each and pDp4 showed ~ 1000, in these species (Table 5 and Additional File 9).
pDp1, pDp2 and pDp3 sequences are dispersed throughout the buffalo genome
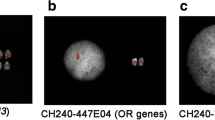
In silico analysis of pDp1, pDp2, pDp3 and pDp4 sequences using reference cattle genome revealed its multiple locations on the cow chromosomes (Figure 5). Probing of pDp1 to buffalo genomic DNA digested with RsaI enzyme detected a strong hybridization signal (Additional File 10) giving rise to a single isomorphic band in all the samples. FISH mapping of pDp1, 2, 3 spectrum red labeled cloned probes showed ubiquitous discernible signals over buffalo metaphase chromosomes (Figures 6 and 7). All the chromosomes showed dispersed pattern with all the three probes used independently. In several chromosomes, signals in the centromeric regions were absent or reduced giving rise to inconsistent pattern. FISH with pDp1 sequence showed more localized signals on the metaphase chromosomes as compared to that detected by pDp2 and pDp3. FISH with spectrum red labeled clone pDp4 probe showed background signals even after washing the slides under high stringent conditions (60°C in 0.1× SSC). This might be due to the short length (339 bp) of the probe pDp4 used coupled with its dispersed genomic organization.
In silico mapping of RsaI sequences, pDp1 (A), pDp2 (B) pDp3 (C) and pDp4 (D) using existing cattle genomic map data (cow build 4 genome database). Note presence of pDp1, pDp2 and pDp4 sequences at multiple locations on cattle chromosomes. Color bar having percent homology of RsaI sequences with cattle genome is shown on right side of the figure.
Full length cDNA sequence of ACOT11 gene in Buffalo
Blast search using reference mRNA sequence revealed RsaI repeats to be part of Acyl-coenzyme A thioesterase 11(ACOT11), V acuolar Protein Sorting 24 (VPS24) and Solute carrier organic anion transporter family member 1A2 (SLCO1A2) genes (Additional File 1) Full length Buffalo ACOT11 cDNA was generated using end point PCR and gene specific (B. taurus ACOT11) primers (Table 3). Assembled cDNA sequence of 2592 base pair fragment lacking poly A tail representing six exons (Additional Files 11 and 12) were deposited in the GenBank (HQ848649 and HQ848650).
Discussion
We have studied four (pDp1, pDp2, pDp3 and pDp4) RsaI fragments from the buffalo genome which are AT rich, though buffalo genome on the whole is GC rich (40.69%, NC_006295). Database searches with the repeat-maskers revealed the presence of several LTR, LINE and SINE element in the four sequences (Table 2). Apparently, SINEs occupy the (G+C)-rich regions while LINEs are mainly located on the (A+T)-rich regions. Reports suggest that very large number of highly truncated insertions of L1 have occurred in the bovine genome [8]. Full length copies of the human L1 contain two open reading frames, ORF1 and 2. ORF1 encodes a DNA binding protein and ORF2 includes endonuclease and reverse transcriptase domains [27]. Presence of partial reverse transcriptase and endonuclease domains in pDp1, pDp2 and pDp4 reported herein led to the hypothesis that RsaI repeats might be related to a novel retrotransposable element.
Interspersed repeats get inserted into a new genomic location through the process of retrotransposition [28, 29]. This is reflected by our FISH results of pDp1, 2 and 3 showing signals on all over the chromosomes with varying intensity. In silico analysis of pDp3 on the reference cattle genome showed fewer distribution suggesting its poor characterization in the cattle genome. This is supported by the fact that using real time PCR, we detected similar copy number of pDp3 in the bovids. Repetitive sequences in centromeric regions are dynamic components, ever prone to mutation, recombination, deletion, and translocation leading eventually to their alterations [30]. Absence of RsaI sequences in the centromere of several chromosomes may be undergoing such events. All the four sequences showed no homology with each other as revealed by clustalW alignment (Additional File 2). Startlingly, these four repeat elements were not detected in any of the non-bovid species. This suggests that irrespective of their origin and biological significance, their evolutions have been confined to limited number of species. Most likely, in non-bovid species, they were not favored evolutionarily and therefore purged slowly and gradually in due course of time. We presume that amplifications of repeat elements could have originated from its discrete blocks. Reintegration of extra chromosomal copies of these repeat elements [31] could have allowed its further dissemination in the genome. However, it is not clear whether all the repeats have similar significance in the buffalo genome. It is likely that these repeats are collectively involved in the evolution and sustenance of bovid chromosomes. Reports suggest that new retrotransposons are conserved within the same group of species [32]. This is corroborated by our Slot blot and PCR results. Real time PCR results showed approximately similar copy numbers for pDp1, 2, 3 and 4 in cattle, goat and sheep genomes as mentioned earlier. The copy number assessment of these repeats in different known and non-descript breeds of buffalo may enable to establish a correlation, if any, towards the delineation of different breeds.
Retrotransposons copies are reportedly involved in the regulation of transcription [33–36]. Presence of RsaI repeats in exonic database of various transcribing genes suggests that these sequences function as parts of mRNA. Since repeat sequences were also present in the introns, it is possible that they are transcribed as pre-mRNA and contribute to the processing of mRNA and splicing. Thus, differential expression of transcripts in somatic tissue and spermatozoa might be under the influence of post transcriptional regulation required for various cellular processes. Presence of transcribing retroelements within the buffalo spermatozoa reported here seems to be first such observation. Studies have shown existence of an RT-dependent mechanism operating in the spermatozoa, responsible for the genesis of new biologically active retrogenes [37]. These retrogenes may be delivered to the embryos during fertilization and propagated subsequently in the tissues of adult individuals [37].
RsaI elements were found to be part of the three functional genes (ACOT11, VPS24 and SLCO1A2) mostly present in 3' UTR. Our work corroborates recent reports that most part of retrotransposons inserts themselves in first and last exons and in untranslated regions (UTRs) [38]. In human, this type of insertion has been shown to create new non-conserved polyadenylation signals [39], influencing the level of gene expression [40]. However, how these insertions affect expression of buffalo transcriptomes is still a matter of speculation.
Conclusions
Buffalo has several known and non-descript breeds of which a few are considered to be superior with respect to productivity and economic return. Whether, RsaI repeat in different breeds of buffalo would show similar organization and expression pattern is not known. However, if informative in breed delineation, these would prove to be useful biomarkers.
Methods
Species sample and DNA extraction
Approximately, 10 ml blood samples from both the sexes of buffalo, goat and sheep were collected from local slaughterhouse, Delhi following the guidelines of institute's Ethical and Biosafety Committee. Cattle blood sample was procured from the owner of the animal. Blood samples of human, fish, bird, rat, jungle cat, bonnet monkey, were available from other projects in the lab. Tiger, Indian rhinoceros and leopard samples were obtained with due permission from the competent authorities of the state and union government of India following strictly the guidelines of the Institute's Ethical and Biosafety Committee. Genomic DNA was extracted according to standard phenol-chloroform procedure [41].
Restriction digestion of buffalo genomic DNA, cloning and sequencing
Approximately, 5 μg of buffalo genomic DNA from both the sexes was digested with RsaI restriction enzyme following supplier's (NEB) specification. Fragments were separated on 1% agarose gel in 1× TBE. Distinct bands within the smear were sliced from the gel, purified and cloned into dephosphorylated pBluescript II SK+ vector (Stratagene, USA), using standard protocol [42]. Approximately, ten clones, representing each fragment, were screened with restriction enzyme Xho 1/Nde 1 for the presence of insert. Slot-blot hybridization was conducted using RsaI fragments as probe labeled by random priming (RediprimeTM II kit, Amersham Pharmacia biotech, USA). Finally, positive clones were selected for sequencing.
In silico analysis
Multiple sequence alignment and Phylogenetic tree construction were carried out using ClustalW program. Blast search were performed with the sequences in GenBank using BLASTN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome version 2.2.18+) with default parameters. Repeats were calculated using Repeat masker http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker. Clustered repeats were found out using http://zlab.bu.edu/repfind/form.html ORFs and amino acid sequence identification was done using http://www.ncbi.nlm.nih.gov/gorf/gorf.html Conserved domain was determined using site http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi Cattle chromosome map was constructed using NCBI Bos taurus genome view.
Buffalo genomic DNA analysis
For cross hybridization studies, approximately 500 ng heat denatured genomic DNA from 14 species each mentioned earlier were slot-blotted onto the nylon membrane (Amersham) along with cloned plasmid as positive control and 2× SSC as negative control following standard protocol [41]. For Southern hybridization, approximately, 4-5 μg of buffalo, cattle, goat and sheep genomic DNA were subjected to restriction digestion using RsaI enzyme following supplier's (NEB) specifications. The digested DNA was resolved on 1.5% agarose gel and transferred onto the nylon membrane (Amersham) following standard protocol [42]. Membranes were rinsed in 2× SSC, dried and UV cross-linked. Blots were hybridized at 60°C overnight with α-32P-dCTP labeled recombinant plasmid (25 ng) using random priming method (rediprimeTM II kit, Amersham Pharmacia biotech, USA). Washing of the membranes was done using standard protocols and signals were recorded by exposure of the blot to X-ray film [41].
PCR
The extent of sequence conservation across bovid genomes was further determined by PCR analysis. Primers were designed from ORF using Primer 3 software (Table 3). 50 ng of genomic DNA from buffalo, cattle, goat and sheep were PCR amplified using reaction conditions 95°C for 5 minutes followed by 35 cycles each consisting 95°C for 1 minute, 60°C for 1 minute and 72°C for 1 minute, and final extension at 72°C for 10 minutes for all the four fragments. Amplified fragments were resolved on 1.5% agarose gel in 1% TAE buffer (Table 3).
RNA isolation and synthesis of cDNA
Total RNA was extracted from testis, kidney, liver, spleen, lung, heart, ovary, brain and sperm using TRIzol (Molecular Research Center, Inc., Cincinnati, OH) following manufacturer's instructions [43, 44]. Tissues were procured from the local slaughterhouse, Delhi. Fresh ejaculated semen samples from buffalo bulls were obtained from the animal farm, Lucknow, U.P., India, strictly following the guidelines of the Institutes Ethical and Biosafety Committee. To check the contamination of mRNA from the cells other than spermatozoa, RNA extractions from the sperms were tested by RT-PCR for both the CDH1 (E-cadherin) GenBank Accession no. AJ400864 and CD45 (tyrosine phosphatase) GenBank Accession no. NM_001002763[45]. Similarly, presence of DNA was ruled out by PCR using β-actin primers, GenBank accession no. DQ661647 (Table 3). Following this, approximately 10 μg of RNA from different tissues and spermatozoa was reverse transcribed into cDNA using commercially available high capacity cDNA RT kit (Applied Biosystems, USA). The success of cDNA synthesis was confirmed by 35 cycles of PCR amplification using buffalo derived β-actin primers.
RT-PCR and Relative expression analysis
Expression analysis of pDp1, pDp2, pDp3 and pDp4 transcripts using 50 ng cDNA from different somatic tissues and spermatozoa of buffalo was done with sequence specific internal primers designed by Primer 3 software (Table 3). PCR conditions were same as that mentioned earlier. β-actin was used as positive control. Relative expression analysis for the four transcripts was done using SYBR Green and Real time PCR Sequence Detection System-7500 (ABI, USA). The primers specific to the sequence were designed using Primer Express Software V2.0 (ABI) (Table 3). Primers for the housekeeping gene GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) GenBank Accession no. XR_083674.1 was used to normalize the values for each sample. The specificity of each primer pair and efficiency of the amplification were tested by assaying serial dilutions of the cDNA. Each reaction was performed in triplicates and the mean value was used for the analyses [46]. The cyclic conditions comprise 50°C for 2 min and 95°C for 10 min, followed by 40 cycles each of 95°C for 10 s and 60°C for 1 min. Each experiment was repeated three times to ensure consistency of the results. The expression level of the desired sequence in different tissues and spermatozoa was calculated in the form of 2-ΔΔCt value http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_042176.pdf[46].
Copy number calculation
Real-time qPCR assays were performed in a 25 μl reaction volume containing 12.5 μl 2× SYBR Green® PCR Master Mix (Applied Biosystems, Foster City, CA, USA), genomic DNA (0.1, 1.0 and 5 ng), forward and reverse primers at final concentration of 100 μM. Copy number estimation for four RsaI fragments in buffalo, cattle, goat and sheep genomes were done using 10 fold dilutions series of recombinant plasmids in the range 30,00,00000 to 3 copies (assuming haploid genome of farm animals =3.3 pg, wt per base pair = 1.096 × 10-21 gm). Reactions were performed in 96-well MicroAmp Optical Reaction Plates (Applied Biosystems) in triplicates using the Real time PCR Sequence Detection System-7000 (ABI, USA) and SYBR Green dye. Primers and assay conditions were similar to those used for Relative expression studies (Table 3). Reaction specificity was confirmed with melting curves analysis. The standard curve was prepared using 10 folds dilution series of the recombinant plasmids and buffalo, cattle, goat and sheep genomic DNA [45, 46].
Chromosome preparation and fluorescence in situ hybridization (FISH)
Approximately, 200 μl of the whole blood from buffalo was cultured for chromosome preparation following standard protocols [46]. FISH was conducted with spectrum red labeled pDp1, pDp2 and pDp3 and pDp4 cloned probes (Abbott Molecular) on the metaphase chromosomes using Nick Translation Kit, Abbott Molecular, (Illinois, USA). Hybridization was carried out in 20 μl volume containing 50% formamide, 10% Dextran sulphate, Cot 1 DNA and 2× SSC, pH 7 for 16 hours at 37°C in a moist chamber. Post hybridization washes were done in 2× SSC at 37°C (low stringent condition) and then at 60°C in 0.1× SSC (under high stringent condition). Slides were counterstained with DAPI, screened under Olympus Fluorescence Microscope (BX51) and images were captured with Olympus U-CMAD-2 CCD camera. Chromosome mapping was done following the International System for Chromosome Nomenclature (ISCND 2000) for Bovids [47].
Generation of full length buffalo ACOT11 mRNA using endpoint PCR
Blast search with pDp1 sequences showed 91% homology with B. taurus ACOT11 gene from nucleotide position 730-1331 encompassing 602 bp. Full length buffalo ACOT11 mRNA was generated using primers designed from B. taurus ACOT 11 (Accession No. NM_001103275). Details of Primer sequences and product size are given in Table 3. PCR amplified products were cloned into and pGEMT-easy vector and sequenced. Finally, buffalo ACOT11 gene sequences were assembled and full length sequence was deposited in the GenBank.
Abbreviations
- CD45:
-
Cluster of differentiation 45
- CDH1:
-
Cadherin-1
- cDNA:
-
complementary Deoxyribonucleic acid
- Ct:
-
Cycle threshold
- DAPI:
-
4',6-diamidino-2-phenylindole
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- mRNA:
-
Messenger ribonucleic acid
- RT-PCR:
-
Reverse transcriptase Polymerase chain reaction
- RTE:
-
Retrotransposable element
- RT-PCR:
-
Reverse Transcriptase-Polymerase Chain Reaction
- TAE:
-
Tris/Acetic acid/EDTA
- TBE:
-
Tris/Borate/EDTA
- UTR:
-
Untranslated region.
References
Charlesworth B, Sniegowski P, Stephan W: The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994, 371: 215-220. 10.1038/371215a0.
Smit AF: The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996, 6: 743-748. 10.1016/S0959-437X(96)80030-X.
Singer MF: SINEs LINEs: highly repeated short long interspersed sequences in mammalian genomes. Cell. 1982, 28: 433-434. 10.1016/0092-8674(82)90194-5.
Rogers J: Origins of repeated DNA. Nature. 1985, 317: 765-766. 10.1038/317765a0.
Nikaido M, Okada N: CetSINEs AREs are not SINEs but are parts of cetartiodactyl L1. Mamm Genome. 2000, 11: 1123-1126. 10.1007/s003350010221.
Kramerov DA, Vassetzky NS: Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005, 247: 165-221.
Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, et al: Genome sequence comparative analysis haplotype structure of the domestic dog. Nature. 2005, 438: 803-819. 10.1038/nature04338.
Malik HS, Eickbush TH: The RTE class of non-LTR retrotransposons is widely distributed in animals is the origin of many SINEs. Mol Biol Evol. 1998, 15: 1123-1134.
Ray DA, Xing J, Salem AH, Batzer MA: SINEs of a nearly perfect character. Syst boil. 2006, 55: 928-935. 10.1080/10635150600865419.
Shedlock AM, Okada N: SINE insertions: powerful tools for molecular systematic. Bioessays. 2000, 22: 148-160. 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z.
Sayah DM, Sokolskaja E, Berthoux L, Luban J: Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004, 430: 569-573. 10.1038/nature02777.
Clark LA, Wahl JM, Rees CA, Murphy KE: Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci USA. 2006, 103: 1376-1381. 10.1073/pnas.0506940103.
Kobayashi S, Goto-Yamamoto N, Hirochika H: Retrotransposon-induced mutations in grape skin color. Science. 2004, 304: 982-10.1126/science.1095011.
Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D: A distal enhancer an ultraconserved exon are derived from a novel retroposon. Nature. 2006, 441: 87-90. 10.1038/nature04696.
Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D: Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007, 104: 18613-18618. 10.1073/pnas.0703637104.
Feschotte C: Transposable elements the evolution of regulatory networks. Nat Rev Genet. 2008, 9: 397-405. 10.1038/nrg2337.
Adelson DL, Raison JM, Edgar RC: Characterization Distribution of Retro-transposons Simple Sequence Repeats in the Bovine Genome. Proc Natl Acad Sci USA. 2009, 106: 12855-12860. 10.1073/pnas.0901282106.
Lev-Maor G, Sorek R, Shomron N, Ast G: The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons. Science. 2003, 300: 1246-1247. 10.1126/science.1085690.
Sorek R, Ast G, Graur D: Alu-containing exons are alternatively spliced. Genome Res. 2002, 12: 1060-1067. 10.1101/gr.229302.
Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D, Ast G: Minimal conditions for exonization of intronic sequences: 5' splice site formation in Alu exons. Mol Cell. 2004, 14: 221-231. 10.1016/S1097-2765(04)00181-9.
Wang W, Kirkness EF: Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005, 15: 1798-1808. 10.1101/gr.3765505.
Cordaux R, Batzer MA: Teaching old dog new tricks: SINEs of canine genomic diversity. Proc Natl Acad Sci USA. 2006, 103: 1157-1158. 10.1073/pnas.0510714103.
Almeida LM, Amaral ME, Silva IT, Silva WA, Riggs PK, Carareto CM: Report of a chimeric origin of transposable elements in a bovine-coding gene. Genet Mol Res. 2008, 7: 107-116. 10.4238/vol7-1gmr371.
Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G: Comparative analysis of transposed element insertion within human mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007, 8: 127-10.1186/gb-2007-8-6-r127.
Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, Pollock DD, Jurka J: Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007, 17: 992-1004. 10.1101/gr.6070707.
Gu W, Ray DA, Walker JA, Barnes EW, Gentles AJ, Samollow PB, Jurka J, Batzer MA, Pollock DD: SINEs evolution genome structure in the opossum. Gene. 2007, 396: 46-58. 10.1016/j.gene.2007.02.028.
Han JS, Szak ST, Boeke JD: Transcriptional disruption by the L1 retrotransposon implications for mammalian transcriptomes. Nature. 2004, 429: 268-274. 10.1038/nature02536.
Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, Devine SE: Recently mobilized transposons in the human chimpanzee genomes. Am J Hum Genet. 2006, 78: 671-679. 10.1086/501028.
Cordaux R, Batzer MA: The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009, 10: 691-703. 10.1038/nrg2640.
Qi L, Friebe B, Gill BS: Complex genome rearrangements reveal evolutionary dynamics of pericentromeric regions in the Triticeae. Genome. 2006, 49: 1628-1639. 10.1139/g06-123.
Hyrien O, Debatisse M, Buttin G, de Saint Vincent BR: The multicopy appearance of a large inverted duplication the sequence at the inversion joint suggest a new model for gene amplification. EMBO J. 1988, 7: 407-417.
Sheikh FG, Mukhopadhyay SS, Gupta P: PstI repeat: a family of short interspersednucleotide element (SINE)-like sequences in the genomes of cattle goat buffalo. Genome. 2002, 45: 44-50. 10.1139/g01-122.
Britten RJ: Mobile elements inserted in the distant past have taken on important functions. Gene. 1997, 205: 177-182. 10.1016/S0378-1119(97)00399-5.
Brosius J: Genomes were forged by massive bombardments with retroelements retrosequences. Genetica. 1999, 107: 209-238. 10.1023/A:1004018519722.
Faulkner GJ, Carninci P: Altruistic functions for selfish DNA. Cell Cycle. 2009, 8: 2895-2900. 10.4161/cc.8.18.9536.
Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest AR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P: The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009, 41: 563-571. 10.1038/ng.368.
Sciamanna I, Vitullo P, Curatolo A, Spadafora C: Retrotransposons reverse transcriptase the genesis of new genetic information. Gene. 2009, 448: 180-186. 10.1016/j.gene.2009.07.011.
Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G: Comparative analysis of transposed element insertion within human mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007, 8: 127-10.1186/gb-2007-8-6-r127.
Gal-Mark N, Schwartz S, Ram O, Eyras E, Ast G: The pivotal roles of TIA proteins in 5' splice-site selection of alu exons across evolution. PLoS Genet. 2009, 5: e1000717-10.1371/journal.pgen.1000717.
Lee JY, Ji Z, Tian B: Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3'-end of genes. Nucleic Acids Res. 2008, 36: 5581-5590. 10.1093/nar/gkn540.
John MV, Ali S: Synthetic DNA-based genetic markers reveal intra-inter-species DNA sequence variability in the Bubalus bubalis related genomes. DNA Cell Biol. 1997, 16: 369-378. 10.1089/dna.1997.16.369.
Sambrook J, Fritschi EF, Maniatis T: Molecular cloning. Cold Spring Harbor Laboratory Press New York. 1989
Srivastava J, Premi S, Kumar S, Ali S: Organization differential expression of the GACA/GATA tagged somatic spermatozoal transcriptomes in Buffalo Bubalus bubalis. BMC Genomics. 2008, 9: 132-10.1186/1471-2164-9-132.
Srivastava J, Premi S, Kumar S, Ali S: Expressional dynamics of minisatellite 33.15 tagged spermatozoal transcriptome in Bubalus bubalis. BMC Genomics. 2009, 10: 303-10.1186/1471-2164-10-303.
Pathak D, Srivastava J, Samad R, Parwez I, Kumar S, Ali S: Genome-wide search of the genes tagged with the consensus of 33.6 repeat loci in buffalo Bubalus bubalis employing minisatellite-associated sequence amplification. Chromosome Res. 2010, 18: 441-458. 10.1007/s10577-010-9132-0.
Pathak D, Srivastava J, Premi S, Tiwari M, Garg LC, Kumar S, Ali S: Chromosomal localization copy number assessment transcriptional status of BamHI repeat fractions in water buffalo Bubalus bubalis. DNA Cell Biol. 2006, 25: 206-214. 10.1089/dna.2006.25.206.
Cribiu EP, Di Berardino D, Di Meo GP, Eggen A, Gallagher DS, Gustavsson I, Hayes H, Iannuzzi L, Popescu CP, Rubes J, Schmutz S, Stranzinger G, Vaiman A, Womack J: International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet Cell Genet. 2001, 92: 283-299. 10.1159/000056917.
Acknowledgements
This work was supported by a DST Grant No.SR/WOSA/LS-92/2005 to DP, DBT Grants No. BT/PR14102/AAQ/01/438/2010 to SA and a core grant from the Department of Biotechnology, Govt. of India to the National Institute of Immunology, New Delhi. SA thanks Alexander Von Humboldt Foundation, Bonn, Germany for equipment donation and Shri Khem Singh Negi for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
DP carried out the experiments and in-silico analysis, interpreted the data, and wrote the manuscript. SA conceived and designed the study, interpreted the results and revised the manuscript critically. All the authors read and approved the final manuscript
Electronic supplementary material
12864_2011_3501_MOESM1_ESM.PDF
Additional file 1:Details of the Blast search. Details of the Blast search of RsaI derived repeat sequences of water buffalo Bubalus Bubalis. (PDF 147 KB)
12864_2011_3501_MOESM2_ESM.PDF
Additional file 2:Details of ClustalW alignment. ClustalW alignment of buffalo derived RsaI element pDp1, pDp2, pDp3 and pDp4, showing each one as separate entity. (PDF 79 KB)
12864_2011_3501_MOESM3_ESM.PDF
Additional file 3:ClustalW alignment with transcribing genes. ClustalW alignment of buffalo RsaI pDp1, pDp2 and pDp4 sequences with Bos taurus transcribing genes (A) ACOT11 (B) VPS24 and (C) SLCO1A2. Sequences highlighted in yellow indicate UTR. RsaI sequences are marked in blue. (PDF 146 KB)
12864_2011_3501_MOESM4_ESM.PDF
Additional file 4:Details of Cross-hybridization studies. Cross-hybridization of RsaI recombinant clones with genomic DNA of different species. Signals were detected only in buffalo, cattle, goat and sheep as shown herein. PC denotes positive control (recombinant plasmids). IDs of the sequences used for hybridization are mentioned on the left. (PDF 130 KB)
12864_2011_3501_MOESM5_ESM.PDF
Additional file 5:Details of Southern blot hybridization across bovids. Representative blots showing distribution of pDp1 (A) and pDp2 (B) in buffalo, cattle, goat and sheep genome by Southern blot hybridization. Note discernible bands of 1331 and 652 bp in these species. (PDF 56 KB)
12864_2011_3501_MOESM6_ESM.PDF
Additional file 6:Details of pDp1 alignment across the species. ClustalW nucleotide alignment of buffalo pDp1, 489 bp ORF sequence, with cattle, goat and sheep sequences. Note the close sequence homology among the bovids. (PDF 42 KB)
12864_2011_3501_MOESM7_ESM.PDF
Additional file 7:Phylogenetic analysis. Phylogram based on percent identity of pDp1, pDp2, pDp3 and pDp4 (A-D) sequence in different species showing close relationship of buffalo with cattle. (PDF 8 KB)
12864_2011_3501_MOESM8_ESM.PDF
Additional file 8:Details of RT PCR. RT-PCR analysis of RsaI repeat sequences using internal primers and cDNA from different somatic tissues and spermatozoa of buffalo, Sequence IDs are indicated on the left and tissues are mentioned on top of the lanes. β-actin was used a positive control. M denotes 100 base pair marker. (PDF 80 KB)
12864_2011_3501_MOESM9_ESM.PDF
Additional file 9:Details of copy number calculation with Real time PCR. Standard curve based on 10 fold dilution series of pDp1, pDp2, pDp3, pDp4 and genomic DNA from buffalo, cattle, goat and sheep showing the amplification plot (a-d) panel (A), corresponding slopes of -3.3 to -3.5, panel (B) and a single dissociation peak, panel (C), substantiating maximum efficiency of the PCR reaction and high specificity of the primers with target DNA. Arrow indicates genomic DNA from buffalo, cattle, goat and sheep. (PDF 343 KB)
12864_2011_3501_MOESM10_ESM.PDF
Additional file 10:Southern hybridization with pDp1 clone. Southern hybridization of Bubalus bubalis RsaI digested genomic DNA with pDp1 clone (A). The strongest isomorphic band corresponds to 1331 bp, indicated by an arrow (B). (PDF 35 KB)
12864_2011_3501_MOESM11_ESM.PDF
Additional file 11:Status of Exons in ACOT11 gene. Pictorial representation showing Bos taurus (A) and Bubalus bubalis ACOT11 gene (B) with their representative exons. Nucleotide position 730 to 1331 indicates region of pDp1 showing 92% homology to Bos taurus ACOT11. Full length sequence of Bubalus bubalis ACOT11 gene lacking poly A tail and exons are given in (C). (PDF 118 KB)
12864_2011_3501_MOESM12_ESM.PDF
Additional file 12:ClustalW alignment of ACOT11 gene. ClustalW alignment of buffalo ACOT11 gene with cattle (NM_001103275.1). Note the high level of sequence homology (92%) between the two species. (PDF 45 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Pathak, D., Ali, S. RsaI repetitive DNA in Buffalo Bubalus bubalis representing retrotransposons, conserved in bovids, are part of the functional genes. BMC Genomics 12, 338 (2011). https://doi.org/10.1186/1471-2164-12-338
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-12-338