Abstract
Background
Bacterial spot of tomato and pepper is caused by four Xanthomonas species and is a major plant disease in warm humid climates. The four species are distinct from each other based on physiological and molecular characteristics. The genome sequence of strain 85-10, a member of one of the species, Xanthomonas euvesicatoria (Xcv) has been previously reported. To determine the relationship of the four species at the genome level and to investigate the molecular basis of their virulence and differing host ranges, draft genomic sequences of members of the other three species were determined and compared to strain 85-10.
Results
We sequenced the genomes of X. vesicatoria (Xv) strain 1111 (ATCC 35937), X. perforans (Xp) strain 91-118 and X. gardneri (Xg) strain 101 (ATCC 19865). The genomes were compared with each other and with the previously sequenced Xcv strain 85-10. In addition, the molecular features were predicted that may be required for pathogenicity including the type III secretion apparatus, type III effectors, other secretion systems, quorum sensing systems, adhesins, extracellular polysaccharide, and lipopolysaccharide determinants. Several novel type III effectors from Xg strain 101 and Xv strain 1111 genomes were computationally identified and their translocation was validated using a reporter gene assay. A homolog to Ax21, the elicitor of XA21-mediated resistance in rice, and a functional Ax21 sulfation system were identified in Xcv. Genes encoding proteins with functions mediated by type II and type IV secretion systems have also been compared, including enzymes involved in cell wall deconstruction, as contributors to pathogenicity.
Conclusions
Comparative genomic analyses revealed considerable diversity among bacterial spot pathogens, providing new insights into differences and similarities that may explain the diverse nature of these strains. Genes specific to pepper pathogens, such as the O-antigen of the lipopolysaccharide cluster, and genes unique to individual strains, such as novel type III effectors and bacteriocin genes, have been identified providing new clues for our understanding of pathogen virulence, aggressiveness, and host preference. These analyses will aid in efforts towards breeding for broad and durable resistance in economically important tomato and pepper cultivars.
Similar content being viewed by others
Background
Bacterial spot disease of tomato and pepper presents a serious agricultural problem worldwide, leading to significant crop losses especially in regions with warm and humid climate. The disease is characterized by necrotic lesions on leaves, sepals and fruits, reducing yield and fruit quality [1]. The disease is caused by a relatively diverse set of bacterial strains within the genus Xanthomonas; strain nomenclature and classification for the strains that infect pepper and tomato have gone through considerable taxonomic revision in recent years. Currently, the pathogens are classified into four distinct pathogen groups (A, B, C, and D) within the genus Xanthomonas. Strains belonging to groups A, B and D infect both tomato and pepper. Group C strains are pathogenic only on tomato [2, 3]. These phenotypically and genotypically distinct strains have different geographic distributions. Strains of group A and B are found worldwide. C strains have been increasingly found in the U.S., Mexico, Brazil, Korea and regions bordering the Indian Ocean, and D group strains are found in the former Yugoslavia, Canada, Costa Rica, U.S, Brazil and regions of the Indian Ocean [4–8]. Three of the four groups except for D were originally described as a single pathovar within Xanthomonas campestris and referred to as X. campestris pv. vesicatoria. The D group consisted of a strain isolated from tomato that had been designated 'Pseudomonas gardneri' for many years [9] although De Ley provided evidence for placement in the genus Xanthomonas[10]. Subsequently all four groups were classified as separate species on the basis of physiological and molecular characteristics as follows: Xanthomonas euvesicatoria (group A), Xanthomonas vesicatoria (group B), Xanthomonas perforans (group C), and Xanthomonas gardneri (group D) [11].
Based on 16S rRNA analysis, X. euvesicatoria strain 85-10 (A group) and X. perforans (C group) together form a monophyletic group, whereas X. vesicatoria (B group) and X. gardneri (D group) cluster together with X. campestris pv. campestris (Xcc) Xcc strain 33913 [11]. Recently, a phylogenetic tree was constructed based on MLST (multi-locus sequence typing) data for A, B, C and D group strains and other xanthomonads [12]. The MLST approach revealed that X. euvesicatoria and X. perforans form a group along with X. citri strain 306 (Xac). X. gardneri is most closely related to X. campestris pv. campestris strains while X. vesicatoria forms a distinct clade [12]. This diversity among the four groups makes the Xanthomonas-tomato/pepper system an excellent example to study pathogen co-evolution, as distinct species have converged on a common host.
While integrated management approaches for control of bacterial spot disease are available, the development of host resistance is more economical and environmentally benign for the control of the disease [13, 14]. Host resistance may also be required to replace the loss of some integrated management tools. Use of copper and streptomycin sprays over the years, for example, has led to the development of resistant strains [5]. At the same time, genetic resistance has been lost due to race shifts in pathogen populations [15–17]. Designing new and possibly durable resistance requires knowledge of pathogenicity factors possessed by the four groups.
Many candidate pathogenicity factors have been identified in strains of Xanthomonas. A number of virulence factors are employed by xanthomonads to gain entry into leaf or fruit tissue, and gain access to nutrients, while simultaneously overcoming or suppressing plant defenses. Different secretion systems and their effectors have been shown to contribute to the virulence of plant pathogens. The type III secretion system (T3SS) encoded by the hrp (Hypersensitive Response and Pathogenicity) gene cluster [18, 19] and type III secreted effectors have been widely studied for their role in hypersensitivity and pathogenicity. Effectors common between strains are believed to be responsible for conserved virulence function and avoidance of host defense. Differences in effector suites have evolved in closely related strains of plant pathogens and strain-specific effectors may help to escape recognition by host-specific defenses [20–25]. Important insights into pathogenicity mechanisms of X. euvesicatoria strain 85-10 (hereafter, Xcv) have been obtained with its genome sequence [26]. Here we report draft genome sequences of type strains of the other three bacterial spot pathogen species: X. vesicatoria strain 1111 (Xv 1111) (ATCC 35937), X. perforans strain 91-118 (Xp 91-118), and X. gardneri strain 101 (Xg 101) (ATCC 19865). We have annotated and analyzed predicted pathogenicity factors in the draft genomes. Additionally, we have investigated differentiation between xanthomonads that might explain differences in disease phenotypes and in host range.
Results and Discussion
Draft genome sequences of Xv strain 1111, Xp strain 91-118 and Xg strain 101 were obtained by combining Roche-454 (pyrosequencing) and Illumina GA2 (Solexa) sequencing data
Initially, we sequenced Xv strain 1111 (ATCC 35937) (hereafter Xv), Xp strain 91-118 (hereafter Xp) and Xg strain 101 (ATCC 19865) (hereafter Xg) by 454 pyrosequencing [27]. De novo assembly using Newbler assembler resulted in 4181, 2360 and 4540 contigs, respectively, for Xv, Xp and Xg, with approximately 10-fold coverage for each strain (Additional file 1: Table S1). Many pathogenicity genes, including type III effectors, existed in the form of fragments given the relatively low coverage of the 454-based assembly. More complete assemblies were obtained using Illumina sequencing [28]. De novo assemblies of around 100-fold coverage were constructed from the Illumina data alone or combined with pre-assembled 454 long reads using CLC Genomic Workbench [29]. Combined 454 and Illumina sequencing produced a much better assembly than either technology alone (Table 1). Therefore, combined assemblies were chosen for all subsequent analyses. The average contig size in the combined 454 and Illumina assemblies was around 18 kb for Xv and Xp, and 10 kb for Xg. The N50 (minimum number of contigs needed to cover 50% of the assembly) values were 37 and 40 for Xv and Xp, respectively, and 83 for Xg indicating that final assemblies consist of a few large contigs allowing reasonably accurate whole genome comparisons.
The three strains were deduced to contain plasmids as evidenced by the presence of genes that are known to be involved in plasmid maintenance (e.g. parB/F genes). We have used adjacency to such genes to infer occurrence of certain other genes on plasmids.
Relationships of the strains to other xanthomonads using whole genome comparisons
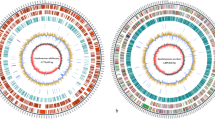
16S rRNA analysis and MLST-based phylogenetic analysis showed the diversity among the four bacterial spot species. We carried out phylogenetic analysis based on orthologous protein-coding genes from draft genomes and reference xanthomonads (Figure 1). Whole genome comparisons were performed using the MUMi index [30] to assess pairwise distance between the draft genomes and available reference Xanthomonas genomes as shown in the phylogenetic tree and the distance matrix (Additional file 2: Fig. S2). Another program, dnadiff, based on nucmer [31] showed the extent of homologies among the shared regions of the genomes by pairwise comparisons (Additional file 3: Table S3). All of the methods yielded consistent results: we were able to ascertain that among the three newly sequenced strains in relationship to the previously sequenced strains, Xp and Xcv form the closest pair, which is in turn closest to Xac. Next, Xg is closest to Xcc, with Xv forming a clade with Xg and the Xcc species group (Figure 1, Additional files S2 and S3).
Maximum likelihood tree based on orthologous genes from xanthomonads and Stenotrophomonas. Concatenated amino acid sequences of the orthologous genes from four bacterial spot pathogen strains along with other sequenced xanthomonads were considered in the analysis. Stenotrophomonas maltophilia (Sm) was used as an outgroup. The evolutionary history was inferred using the Maximum likelihood method. The tree is drawn to scale, with branch lengths corresponding to the evolutionary distances. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site.
Four xanthomonads show variation in the organization of the type III secretion gene clusters
Annotation of the respective type III secretion gene clusters, or hrp genes showed that Xp has an almost identical and syntenic hrp cluster to that of Xcv (Figure 2). The most notable difference is that hpaG and hpaF encode the fusion protein XopAE in Xp, while they are present as separate genes in Xcv. Adjacent hypothetical protein XCV0410 (126 amino acid protein) is absent from Xp. Xv and Xg show greater similarity to the core hrp cluster genes of Xcc than to that of Xcv. Xv and Xg contain hrpW associated with the hrp cluster as in Xcc. Additionally, xopD in Xv and Xg is not associated with the hrp cluster as in Xcc (referred to as psv in Xcc). PsvA shows 74% and 84% sequence identity to the respective homologs from Xv and Xg. XopA (hpa1) from Xcv seems to be absent from Xv and Xg. Interestingly, we found a novel candidate effector gene (named xopZ2) upstream of hrpW in Xv and Xg (See below, Additional file 4: Fig. S4). Finally, the hrp-associated effector xopF1 is conserved and intact in all four tomato and pepper pathogens.
Comparison of type III secretion system cluster, its associated type III effector genes and helper genes of three draft genomes with already sequenced xanthomonads. Type III secretion gene clusters in five strains are shown. Boxes of the same colour indicate orthologous genes. Genes of special interest discussed in the paper are labeled. Xp has near identical hrp cluster as Xcv; Xv and Xg contain mosaic hrp cluster with organization and gene content similar to Xcc, but associated effectors are similar to Xcv along with novel effector gene associated with the cluster.
A reporter gene assay confirms translocation of novel type III effectors
We identified and annotated T3SS effectors from the three newly sequenced xanthomonads (See Methods). Several candidate effectors, which had not yet been experimentally confirmed in xanthomonads, and candidate effectors with plausible translocation motifs were identified (Tables 2, 3, and 4). Corroborative evidence for T3SS-mediated translocation of the candidate effectors was assessed by constructing fusion genes with the C-terminal end of AvrBs2 coding sequence (avrBs262-574aa) in a race 6 strain of X. euvesicatoria. Translocation was measured in pepper cv. ECW 20R, containing the resistance gene Bs2 (Additional file 4: Fig. S4). Genes xopAO, xopG, xopAM, and XGA_0724 (belonging to the avrBs1 class of effectors), of which homologs were previously found in Pseudomonas species, were demonstrated to direct AvrBs2-specific hypersensitive reactions in ECW 20R (Tables 3, Table 4, Additional file 4: Fig. S4). Another candidate effector gene xopZ2, associated with the hrp clusters in Xv and Xg (Figure 2), was also functional in the AvrBs2-based assay. Thus, we identified five effectors (xopAO, xopG, xopAM, xopZ2, XGA_0724) that have not been previously recognized in Xanthomonas and showed their functionality.
Core effectors among four xanthomonads give insight into infection strategies of the pathogen
Comparing the draft genome sequences of the three xanthomonads with that of Xcv allowed us to identify the core effectors conserved in all four strains as well as strain-specific effectors (Tables 2, 3, and 4).
At least 11 effector genes form a core set of common effectors for xanthomonads infecting tomato and pepper (Table 2). Of these 11, eight effector genes (avrBs2, xopK, xopL, xopN, xopQ, xopR, xopX and xopZ) were found to be conserved in all sequenced xanthomonads including the three draft genomes presented here with the exceptions of X. albilineans and X. campestris pv. armoraciae. These genes might be necessary for maintaining pathogenicity of these xanthomonads in a wide range of host plants. XopN has been reported to suppress PAMP (pathogen-associated molecular pattern)-triggered immunity by interacting with tomato TARK1 and TFT1 [32]. XopF1 is conserved in tomato and pepper xanthomonads. Although a homolog of xopF1 is found in Xcc, the respective gene is truncated [34]. Hence, xopF1 is a potential pathogenicity determinant in tomato. A xopF1 deletion mutant of Xcv did not show any difference in virulence when compared to wild type Xcv on the susceptible cultivar of pepper ECW, suggesting XopF1 is not the lone factor for pathogenicity of Xcv on pepper [33]. Another effector gene, xopD, is associated with the hrp gene cluster in Xcv and Xp. However, xopD appears to have translocated to another location in the genome in case of Xg, Xv and Xcc strains. XopD is annotated as "Psv virulence protein" in Xcc genome [34] and has been shown to be a chimeric protein sharing a C terminus with XopD from Xcv[35]. Although xopD homologs from Xv and Xg are syntenic with the psv gene in Xcc, Xv and Xg have intact full-length copies of xopD as in Xcv, indicating that the xopD could be another effector exclusive to the tomato pathogens and a possible pathogenicity determinant in tomato. XopD has been shown to enhance pathogen survival in tomato leaves by delaying symptom development [36]. Two tandem copies of xopX are found in Xg. However, one gene in Xg appears to be inactive due to a frameshift mutation. In Xp, the two copies of xopX are found in different locations in the genome with neighboring genes, including chaperone gene groEL, which is also duplicated. Orthologs of xopZ are also found in all four xanthomonads, with 82% identity for Xcv and Xp and 35% identity for Xg and Xv. Apart from low sequence identity in Xv and Xg, gene-specific rearrangements appear to have occurred within each ortholog. We propose that the overall low amino acid relatedness (pairwise sequence identities below 50%) of this effector in Xv and Xg warrants assigning the proteins to a new family within the xopZ class, named xopZ2, while the orthologs from Xcv and Xp belong to family of xopZ1 as originally described in Xoo and as supported by pairwise sequence identities of at least 60% (see above, Figure 2, Table 4).
Effectors unique to Xp might be responsible for restricting growth on pepper
Xp is pathogenic only on tomato. The avirulence gene, avrXv3, present in Xp, was previously shown to elicit an hypersensitive response (HR) in pepper cv. ECW [37]. An avrXv3 knockout mutant of Xp is not virulent in pepper cv. ECW indicating that other factors are associated with host specificity. Comparing effector repertoires of the pepper pathogens Xg, Xcv, and Xv with Xp may provide clues to the factors that are responsible for reduced virulence (Table 4). Besides avrXv3, the only effectors present in Xp and absent or inactive in Xg, Xv and Xcv are xopC2, xopAE and xopJ4 (avrXv4) (Table 3). The gene avrXv4 is absent from other sequenced xanthomonads and shows gene-for-gene interaction with the Xv4 resistance gene from the wild tomato relative Solanum pennellii but does not contribute to restricted growth of Xp on pepper [38]. The effector xopC2 is a homolog of the effector rsp1239 from Ralstonia solanacearum GMI1000 and xopAE encodes an LRR protein with homology to the R. solanacearum effector PopC. Both genes, xopC2 and xopAE, are truncated in Xcv. Therefore, these two effectors may trigger immunity in pepper. Interestingly, Xp contains a paralog of xopP. The two copies are found next to each other in the genome and share 75% identity at the amino acid level. The second copy is next to the candidate effector xopC2, which is unique to Xp among tomato and pepper pathogens. Effectors xopC2 and xopP may both act to restrict growth in pepper. Moreover, there are at least two effectors, xopE2 and xopG, present in the pepper pathogens Xcv, Xv and Xg but absent from Xp. These effectors may be essential pathogenicity factors in pepper.
Species-specific effectors
Xv possesses two unique effector genes, xopAG (avrGf1) and xopAI (Table 3). A phylogenetic analysis of xopAG showed that xopAG from Xv is closely related to xopAG from X. citri Aw , which has been shown to be responsible for causing an HR on grapefruit [39]. XopAI is a chimeric protein, which contains a conserved myristoylation motif at its N terminus, like XopJ1. This effector class also includes the homolog XAC3230 from Xac as well as XAUB_26830 and XAUC_23780 from X. fuscans subsp. aurantifolii strains B and C, respectively [25]. The presence of transposons and phage elements in close proximity helps to explain the evolution of this novel effector in Xac by terminal reassortment [35]. Xv also contains effector gene avrBsT, which is responsible for the hypersensitive response on pepper. Loss of the plasmid containing avrBsT in Xcv strain 75-3 allows the strain to cause disease on pepper [40].
Xg contains at least two effectors, avrHah1 (an avrBs3-like effector gene) and xopB as does Xcv, and share sequence identity of 82% and 86% respectively to the corresponding effectors of Xcv. However, AvrHah1 appears to specify a different phenotype when compared to avrBs3 from Xcv. AvrHah1 was shown to be responsible for increased watersoaking on pepper ECW-50R and 60R, whereas Xcv strains carrying avrBs3 show a phenotype that consists of small raised fleck lesions on pepper [41]. Another effector gene, xopB, has a PIP box at the 5' end in Xcv, whereas the homolog in Xg does not contain a PIP box. Neighboring genes to xopB in the respective strains are completely different between genomes, suggesting lack of synteny between the two species in this region (Table 4). XopB from Xg is 92% identical at the amino acid level to the homolog in Xcv. Deletion mutants of xopB from Xcv did not show any difference in virulence, indicating it does not contribute significantly to virulence [42]. However, xopB may contribute to virulence in Xg. We also identified eight effector genes that are unique to Xcv (Table 3). With the exception of xopAA (early chlorosis factor), all of these genes belong to regions of low GC content compared to average genome GC content (64.75%): avrBs1 (42%), xopC1 (48%), xopJ1 (xopJ) (57%), xopJ3 (avrRxv) (52%), xopO (52%), xopAJ (avrRxo1) (51%).
Few effectors are shared among phylogenetically related group strains
Although Xp and Xcv, and Xv and Xg form distinct phylogenetic groups (Figure 1), relatively few effectors are shared between these species. For Xp and Xcv, they share at least six effectors - xopE1, xopF2, xopP, xopV, xopAK, xopAP, which are absent from the other two genomes (Table 4). Xv and Xg appear to be most closely related to strains of X. campestris pv. campestris, and this relationship is reflected in the suite of effector genes. In fact, Xg and Xv share four effector genes with Xcc, namely, xopAM, avrXccA1, hrpW and xopZ2, with the caveat that hrpW and avrXccA1 may not function as intracellular effectors (Table 4). Furthermore, the genomic regions containing these genes are syntenic in Xg, Xv and Xcc.
X. gardneri shows evidence of effector acquisition by horizontal gene transfer
Effector homologs of avrA, hopAS1 and avrRpm1 from P. syringae pv. tomato T1 and P. syringae pv. syringae B728a are found in Xg with 79%, 41% and 61% identity at the amino acid level, respectively (Table 3, Additional file 4: Fig. S4). Other X. gardneri strains also contain these effectors based on PCR screening (data not shown). These three effectors, XGA_0724 (belonging to avrBs1 class), XGA_0764/XGA_0765 (xopAS) and XGA_1250 (xopAO), are unique to X. gardneri. The C terminal region of XGA_0724 shows 53% identity to avrBs1 from Xcv. Hence according to the Xanthomonas effector nomenclature [24], XGA_0724 from Xg was placed under the class avrBs1. XGA_0764/XGA_0765 and XGA_1250 have not yet been reported to be found in xanthomonads and were assigned to new classes xopAS and xopAO. X. gardneri strains have been found to be associated with tomato and have a lower optimum temperature for disease development similar to that of pathovars of Pseudomonas syringae[43]. A high score by Alien_hunter analysis [44], along with very low GC content (45% for XGA_0724 and 48% for XGA_01250, 59% for XGA_0764/XGA_0765) and the proximity of mobile genetic elements provides evidence for horizontal gene transfer (Additional file 5: Table S5). Effector xopAS appears to be separated into two ORFs XGA_0764 and XGA_0765 by internal stop codon. The functionality of effector xopAS needs to be confirmed by in planta reporter gene assay. AvrA of P. syringae pv. tomato PT23 was shown to contribute to virulence on tomato plants [45]. Acquisition of XGA_0724 by Xg might have conferred increased virulence on tomato. AvrRpm1 from P. syringae pv. syringae possesses a myristoylation motif, which is absent from homologs in Xg. This modification in Xg might have been acquired to escape host recognition. Another candidate effector gene, xopAQ, in Xg is found 68 bps downstream of a perfect PIP box. The gene shows 65% identity at the amino acid level to rip6/11, a novel effector from R. solanacearum RS1000 [46].
All four xanthomonads contain Ax21 coding gene but only Xcv contains a functional sulfation gene
The ax21 (activator of XA21-mediated immunity) gene is conserved among Xanthomonas species and is predicted to encode a type I-secreted protein that may serve as a quorum sensing signaling molecule [47]. A 17-amino acid sulfated peptide from the N-terminal region of Xanthomonas oryzae pv. oryzae (Xoo) Ax21 (axYS22) was shown to bind and activate the XA21 receptor kinase from rice, demonstrating that Ax21 is a conserved PAMP that can activate plant immune signaling [48]. The ax21 gene is present in Xcv (93% identity with Xoo PXO99 protein), Xp (94%), Xv (91%), and Xg (88%). The axYS22 peptide is 100% conserved in Xcv, Xp and Xv, while in Xg there is a change from leucine to isoleucine at residue 20; this is unlikely to alter the activity of the peptide, since changing this residue to alanine had no effect on recognition by XA21 [48].
Recognition of axYS22 by the XA21 receptor requires sulfation of tyrosine 22, which requires the putative sulfotransferase RaxST. In contrast to ax21, the raxST gene is more variable in these genomes, which is consistent with a report of sequence differences in this gene among Xoo strains [49]. Furthermore, in Xp, there is a single-nucleotide insertion at position 65, causing a frameshift mutation. The Xv and Xg genomes do not contain raxST; therefore, the ax21 gene products may be nonfunctional in these strains. These findings have implications for the further study of the role of Ax21 in quorum sensing and virulence, as well as for the usefulness of the XA21 receptor to confer resistance to xanthomonads in crop plants.
Two type II secretion systems are conserved in all four Xanthomonas genomes
Most cell-wall degrading enzymes, such as cellulases, polygalacturonases, xylanases, and proteases, are secreted by a type II secretion system (T2SS). The Xps T2SS, present in all xanthomonads, has been studied for its contribution to virulence in Xcc and Xoo[50, 51]. Another T2SS cluster, known as the Xcs system, is found only in certain species of Xanthomonas, e.g. Xcc, Xac, and Xcv. The Xps system secretes xylanases and proteases and is under control of hrpG and hrpX[52], indicating differential regulation. Both Xps and Xcs systems are present in all three draft genomes.
Xanthomonads possess diverse repertoires of cell-wall degrading enzymes, which are present in diverse genomic arrangement patterns
Each species of Xanthomonas has its own collection of genes encoding endoxylanases, endoglucanases, and pectate lyases which contribute to cell wall deconstruction during pathogenesis. We have compared these repertoires from the three draft genomes and other xanthomonads as detailed in Table 5. The genes are designated for different families of glycosyl hydrolases (GH) and polysaccharide lyases (PL) that include the enzymes that cleave glycosidic bonds in the structural polysaccharides of plant cell walls.
Genes encoding secreted endoxylanases regulated by the xps genes have been described for their contributions to virulence, including XCV0965 [52] encoding GH30 endoxyalanase. The GH30 family catalyses the cleavage of methylglucuronoxylans in the cell walls of monocots and dicots at a β-1,4-xylosidic bond penultimate to one linking the xylose residue that is substituted by an α-1,2-linked 4-O-methylglucuronate residue [53, 54]. Such an enzyme secreted by Erwinia chrysanthemi generates oligosaccharides that are not assimilated for growth, suggesting a function in which it contributes to cell wall deconstruction for access to pectates for growth substrate [53]. It is interesting to note the orthologous genes encoding GH30 enzymes are absent in Xg and Xv, with a truncated xyn30 gene in Xac. On the basis of sequence homology, xyn30 genes may also contribute to virulence in Xoo, Xcc and Xp.
The more common GH10 endoxylanases, which occur in several bacterial and fungal phyla, have been implicated in the virulence of plant pathogenic bacteria and fungi [55, 56]. In Xoo, deletion of the gene encoding a GH10 xyn10B resulted in diminished virulence [57]. All sequenced Xanthomonas genomes contain either two or three copies of xyn10 genes, all of which are within a gene cluster that may comprise a single operon (Figure 3). The GH10 endoxylanases are the best studied of all of the xylanases, and structure/function relationships may be inferred on the basis of gene sequence. The action of these enzymes on glucuronoxylans generates xylotriose, xylobiose, and small amounts of xylose that generally serve as substrates for growth. Also generated is methylglucuronoxylotriose, that is formed to the extent that xylose residues in the β-1,4 xylan backbone are substituted with α-1,2-linked 4-O-methylglucuronate residues [58].
Xylanase cluster organization. Three types of cluster organizations can be found within xanthomonads. A) Found in Xac, Xcv and Xp containing three endoxylanase genes xyn10A, xyn10B and xyn10C; B) Found in Xcc, Xv and Xg containing two endoxylanases xyn10A and xyn10C; and C) Found in Xoo containing xyn10A and xyn10B within endoxylanase operon.
An adjacent gene cluster in an opposite orientation contains agu67 gene encoding a GH67 α-glucuronidase that serves to catayze the removal of 4-O-methylglucuronate from the reducing terminus of methylglucuronoxylotriose. This activity provides a synergistic function to the overall xylanolytic process to generate xylotriose, which is converted to xylose by xylanases and xylosidases for complete metabolism [59]. The coregulation of operons encoding XynB and Agu67 enzymes occurs as a logical condition to coordinate expression of genes that encode these and additional enzymes that collectively process glucuronxylans and glucuronoarabinoxylans for complete metabolism. The accessory enzymes and transporters necessary for the function of these enzymes are embedded within these operons in Gram positive bacteria [60–62] and share similarities noted here with Xanthomonas spp.. These include the genes encoding two glycohydrolases, a β-xylosidase and an α-L-arabinofuranosidase. Also included in this cluster are genes encoding enzymes for intracellular metabolism of glucuronate and xylose, including glucuronate isomerase; xylulose isomerase; D-mannonate dehydratase; and D-mannonate oxidoreductase. Genes encoding mannitol dehydrogenase and the hexuronate transporter, as well as the TonB-dependent receptor and LacI transcriptional regulator, flank these two operons.
The arrangement and content of xylanolytic enzymes differentiate Xanthomonas species into three groups (Figure 3). Here, we propose a common nomenclature for xylanases, the genes for which have been annotated in the sequenced genomes. Members of the first group are Xac, Xcv and Xp in which all three genes encoding GH10 endoxylanases (xyn10A, xyn10B and xyn10C) are present, and with additional genes further downstream in this cluster. Members of the second group are Xcc, Xv and Xg in which genes encoding two of the three endoxylanases are present (xyn10A and xyn10C) and where one or more of the the downstream genes are absent. Xoo strains represent a third group in which a different set of two endoxylanase encoding genes are present (xyn10A and xyn10B) and where the β-galactosidase and gluconolactonase genes flanking xyn10C are absent. It is noteworthy that the organization of genes in the cluster encoding the α-glucuronidase is conserved across Xanthomonas species.
Genes involved in several Type IV secretion systems are present in genomes and plasmids
Like Xcv, the tomato pathogens, Xg, Xv and Xp, also appear to contain more than one copy of a type IV secretion system (T4SS) cluster (Figure 4A, B). Two T4SS clusters (Vir and Dot/Icm type) are present in Xcv, and genes belonging to both of these systems are found on plasmids [26]. The Dot/Icm type system is absent from Xv, Xp and Xg.
In Xv and Xp, genes for one T4SS are on a plasmid and the second one on the chromosome while in Xg, two T4SS gene clusters are on a plasmid and one is on the chromosome. The two T4SS clusters on plasmids of Xg do not show any similarity to the genes for T4SS in Xac, Xcv, Xcc and Xoo. Of the two T4SS clusters in Xg, one is also found in Xv and Xp. This cluster appears to be exclusive to these three tomato pathogens (Figure 4A). The genes belonging to this cluster show low (30-45%) identity to the T4SS clusters from Ralstonia, Burkholderia, Bradyrhizobium, and Stenotrophomonas maltophilia. The other cluster from Xg, which is absent from Xv and Xp, shows very high identity (98%) and synteny to the T4SS cluster of Burkholderia multivorans and around 89% identity to a T4SS cluster of Acidovorax avenae subsp. citrulli (Figure 4B).
Apart from the plasmid borne T4SS genes, Xcv also contains a portion of a type IV system cluster on the chromosome and consists of VirB6, VirB8, VirB9, VirD4 genes. This chromosomal cluster is flanked by a transposon element (IS1477) that might indicate its horizontal gene transfer. Xp, Xg and Xv genomes contain a complete chromosomal T4SS cluster showing high identity to the T4SS chromosomal clusters from Xcc (Figure 4C).
Type V secreted adhesins function in synergism during pathogenesis
Different adhesins have been shown to function at different stages of the infection process starting with attachment, entry, later survival inside host tissue and colonization by promoting virulence [63, 64]. FhaB hemagglutinin, important for leaf attachment, survival inside plant tissue and biofilm formation, is present in all four tomato pathogens. In Xcv, fhaB is divided into two separate open reading frames, XCV1860 and XCV1861, with the two-partner secretion domains being present in XCV1860. Sequence alignment indicates that fhaB is possibly inactivated in Xcv by the internal stop codon that separates XCV1860 from XCV1861. In the case of Xoo PXO99A, the Xanthomonas adhesin-like proteins XadA and XadB promote virulence by enhancing colonization of the leaf surface and leaf entry through hydathode [64]. As in Xcv and Xac, Xp encodes two copies of xadA, while Xv and Xg possess a single ortholog of xadA as does Xcc. YapH and the type IV pilus protein PilQ were shown to be involved in virulence in Xoo during later stages of growth and migration in xylem vessels. In Xcv, Xc, and Xoo KACC, two copies of yapH are present. There are two pilQ orthologs in Xcv and only one in other sequenced xanthomonads. Next to the fhaB and fhaC adhesin genes, hms operon is present in the genomes of xanthomonads, the homologs of which are pga operon genes in E. coli involved in biofilm formation [65].
Type VI secretion system is present in Xcv, Xv and Xp
Type VI secretion system (T6SS) has been shown recently to contribute to host pathogen interactions during pathogenesis in Vibrio cholerae, Burkholderia pseudomallei and Pseudomonas aeruginosa. Hcp (Haemolysin-coregulated protein) and Vgr (valine-glycine repeats) proteins are exported by the T6SS [66]. T6SS clusters can be assigned to three different types in xanthomonads (Table 6). Xcv and Xp possess two types of T6SSs (type 1 and 3); whereas Xv contains only a single type of T6SS, type 3. As in Xcc, there is no T6SS cluster in Xg (Table 6, Additional file 6: Table S6).
LPS locus displays remarkable variation in sequence and number of coding genes and shows host specific variation
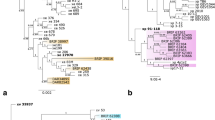
The lipopolysaccharide (LPS) biosynthesis cluster has been studied in detail in Xcc[67], which comprises three regions; region 1 from wxcA to wxcE involved in biosynthesis of water soluble LPS antigen; region 2 (gmd, rmd) coding for LPS core genes; and region 3 from wxcK to wxcO coding for enzymes for modification of nucleotide sugars and sugar translocation systems. This LPS biosynthesis locus is positioned between highly conserved housekeeping genes, namely cystathionine gamma lyase (metB) and electron transport flavoprotein (etfA), as reported in other xanthomonads [68]. Comparison of this cluster from draft genomes to the already sequenced xanthomonads revealed high variability in the number of genes and their sequences. Xv and Xg have an identical type of LPS gene cluster of 17.7 kb encoding 14 open reading frames (Figure 5A) which is similar in organization and sequence identity to the LPS locus from Xcc strains. Interestingly, Xg and Xv also contain two glycosyl transferases involved in synthesis of xylosylated polyrhamnan as seen in Xcc[69], in contrast to glycosyl transferases (wbdA1, wbdA2) involved in synthesis of polymannan in Xcv[26]. This suggests that basic structure of O-antigen in Xg and Xv is similar to Xcc. The three tomato/pepper pathogens Xcv, Xv and Xg have retained an ancestral type of LPS gene cluster (Figure 5A and 5B). On the other hand, Xp has acquired a novel LPS gene cluster during the course of evolution and is completely different in sequence and number of genes that are encoded. In Xp, this LPS locus is 17.3 kb long and encodes 12 ORFs, all of which are absent in the corresponding genomic region of Xcv, Xv or Xg. Also the first five ORFs flanking the metB side of the LPS locus in Xp (Figure 5A, ORFs colored in red) showed very low or no identity to region 1 of the LPS locus in the other xanthomonads. However, these ORFs still belong to the same Pfam families [70] that are usually present in this region, for example, ABC transporters and glycosyl transferases. The second half of the LPS cluster flanking etfA side encodes six ORFs, which are homologs of the LPS cluster genes from Xac, Xcm and Xoo. Phylogenetic insight based on conserved metB and etfA genes that flank the LPS locus suggest that the ancestor of all the Xanthomonas pathogens of pepper and tomato studied in this paper had the same LPS gene cluster, however putative horizontal gene transfer events at this locus have led to the acquisition of a novel LPS gene cluster in Xp (Figure 5B). Alien_hunter analysis also supports this acquisition with a high score showing this region to belong to an anomalous region (Additional file 5: Table S5). This event might have played a major role in changing the specificity of Xp towards tomato and its dominance over its relative(s) as reported previously [71], similar to variant epidemic strain of Vibrio cholerae, reported to be a major reason for its emergence and cholera outbreak during the 1990's in the Indian subcontinent [72]. Identity in terms of sequences and gene organization among pepper pathogens and absence of those genes from X. perforans and a novel LPS cluster in the tomato pathogen X. perforans suggest a role of this cluster in host specific variation.
The Structure and phylogeny of the LPS cluster. A) Schematic comparison of LPS gene clusters described in the present study. Genes conserved in different strains are given identical color. Genes specific to individual strains are given unique color. "Hpo pro" indicates an ORF encoding for a hypothetical protein. The red color-coded genes in Xp genes are absent in any of the sequenced xanthomonads. B) Phylogenetic tree based on conserved metB and etfA genes that flank the variable LPS locus. Strains abbreviations are as in the main text. Arrow indicates the horizontal gene transfer event in the lineage that gave rise to Xp.
Analysis of DSF cell-cell signaling system
RpfC/RpfG are two-component signaling factors and are involved in DSF (diffusible signal factor) cell-cell signaling [73–76], known to co-ordinate virulence and biofilm gene expression. The genomes of Xv, Xp, and Xg carry an rpf (r egulation of p athogenicity f actors) gene cluster (Table 7) that is found in all xanthomonads and which encodes components governing the synthesis and perception of the signal molecule DSF [74, 75]. The Rpf of the DSF system regulates the synthesis of virulence factors and biofilm formation and is required for the full virulence of Xcc, Xac, Xoc, and Xoo[77–81]. RpfF is responsible for the synthesis of DSF, whereas, RpfC and RpfG are implicated in DSF perception and signal transduction [73–76]. RpfC is a complex sensor kinase, whereas RpfG is a response regulator with a CheY-like receiver domain that is attached to an HD-GYP domain. HD-GYP domains act in degradation of the second messenger cyclic di-GMP [82]. In addition to genes encoding these products, Xg and Xp have rpfH, which encodes a membrane protein related to the sensory input domain RpfC but whose function is unknown. Xv contains rpfH but with an internal stop codon, whereas functional rpfH is present in Xcv and Xcc, and totally absent in Xac and Xoo.
Cyclic di-GMP signaling
Cyclic di-GMP is a second messenger known to regulate a range of functions in diverse bacteria, including the virulence of animal and plant pathogens [83–85]. The cellular level of cyclic di-GMP is controlled by a balance between synthesis by GGDEF domain diguanylate cyclases and degradation by HD-GYP or EAL domain phosphodiesterases. GGDEF, EAL and HD-GYP domains are largely found in combination with other signaling domains, suggesting that their activities in cyclic di-GMP turnover can be modulated by environmental cues. A number of proteins involved in cyclic di-GMP signaling have been implicated in virulence of Xcc[86, 87]. The genome of Xcv encodes 3 proteins with an HD-GYP domain and 33 proteins with GGDEF and/or EAL domains. As in other Xanthomonas spp., the HD-GYP domain proteins are completely conserved in Xcv, Xv, Xg and Xp. There is also almost complete conservation of GGDEF/EAL domain proteins between Xcv and three draft genomes, although Xv has no ortholog of XCV1982 (Additional file 7: Table S7). In addition, the EAL domain protein (XCVd0150) encoded on a plasmid in Xcv is absent in the other strains.
Copper resistance (cop) genes are present in Xv and copper homeostasis (coh) genes are present in all strains
Among the Xcv, Xv, Xp and Xg strains sequenced, Xv is the only one resistant to copper and the only strain harboring a set of plasmid borne genes, namely copL, copA, copB, copM, copG, copC, copD, and copF that are also present in copper resistant strains of Xac (unpublished data/Behlau, F. personal communication) and S. maltophilia[88]. Genes copA and copB have been previously annotated as copper resistance related genes for many different xanthomonad genomes including Xoo, Xoc, Xcv, Xac and Xcc. Homologs of these genes are also present in Xv, Xg and Xp and are located on the chromosome. Additionally, upstream of copA on the chromosome of all strains, there is an ORF that shares homology with plasmid copL. In contrast to what has been published, chromosomal copA and copB are not responsible for copper resistance but likely for copper homeostasis and/or tolerance. While strains harboring the plasmid-borne cop genes, like in Xv, are resistant to copper and can grow on MGY agar (manitol-glutamate yeast agar) amended with up to 400 mg L-1 of copper sulfate pentahydrate, strains that have only the chromosomal cop genes as for Xcv, Xp and Xg, are sensitive to copper and can only grow on media amended up to 75 mg L-1 of copper. Nucleotide sequence of plasmid cop genes in Xv are 98% similar to the ones found in Xac and Stenotrophomonas, whereas chromosomal copLAB from Xv is 83% identical to homolog ORFs in Xcv, Xg and Xp. When copL, copA and copB genes from Xv located on the plasmid are compared to the homologs on the chromosome of the same strain, the identity of nucleotide sequences is 27, 73, and 65%, respectively. To avoid further confusion or misinterpretation, we suggest that the nomenclature of the chromosomal copL, copA and copB genes in xanthomonads should be changed to cohL, cohA and cohB, respectively, referring to copper homeostasis genes. New nomenclature has been adopted in the annotation of the draft genomes.
Genes unique to X. perforans as compared to pepper pathogens give clues to its predominance over Xcv in the field and host specificity
Thirteen gene clusters were found to be specific to the tomato pathogen Xp when compared to the other three strains (Additional file 8: Table S8). A part of the clusters are syntenic to the genomic regions specific to the three pepper pathogens, suggesting the replacement of these genomic regions from pepper pathogens in correspond to these region in Xp. These replaced regions in Xp might provide potential candidates for host range determinants. Most notable among these regions was the LPS cluster genes (See above). Other such regions include the avirulence genes avrXv3 and avrXv4, a TIR-like domain containing protein, oxidoreductases, and bacteriocin-like proteins that were not found in any other sequenced xanthomonads. Importance of bacteriocin-like genes in Xp has already been studied for its predominance in the field over T1 strains [89, 90]. Alien_hunter analysis showed that the bacteriocin BCN-A region belongs to an anomalous region indicating possible horizontal gene transfer of this region (Additional file 5: Table S5).
Pepper pathogenicity/aggressiveness factors increased in planta growth of Xp
Comparison of proteomes of Xv, Xg, Xcv against Xp showed 68 genes exclusive to pepper pathogens which might be candidate virulence factors on pepper (Additional file 9: Table S9). These include 16 genes with known function, 35 coding for mobile genetic elements, and 17 genes with unknown function/hypothetical proteins. Out of the 16 genes with known function, xopG was confirmed to be a type III effector using the avrBs2 reporter gene assay and 6 genes belong to the LPS biosynthesis gene cluster. These 16 genes were searched against already sequenced genomes of Xac, Xcc and Xoo. The wxcO gene, which codes for O-antigen, has been identified to be a virulence factor in the X. fuscans - bean pathosystem by subtractive hybridization [91]. Three genes, XCV1298, XCV1839 and wxcO, were initially selected for the verification of their contribution to virulence in pepper. Individual genes along with their promoter regions were cloned into pLAFR3 and conjugated individually and in combination into X. perforans ME24 (91-118ΔavrXv3), which no longer elicits an HR in pepper. However, in planta growth of ME24 is more similar to that of an avirulent strain than the virulent pepper strain TED3 race 6. ME24 transconjugants carrying wxcO and XCV1839 in combination showed increased in planta growth and also comparatively increased number of lesions on pepper cv. ECW when compared to ME24 revealing that these two genes play in fact a role in pepper pathogenicity (Figure 6).
Genes specific to Xg as compared to other tomato/pepper pathogens may explain its aggressive nature on tomato and pepper
Comparison of genes from Xg against Xcv, Xp and Xv genes showed the presence of 625 genes specific to Xg (Additional file 10: Table S10). These include four type III effectors (avrBs1 member, xopAO, avrHah1, xopAQ), twenty-one genes belonging to the unique type IV secretion system cluster and associated genes. These genes can be speculated to contribute to the aggressive nature of Xg strains on tomato and pepper. Xg also contains a unique beta xylosidase not present in any other xanthomonads. Type II secreted beta xylosidase has been studied for its role in plant cell wall digestion. Moreover, Xg contains XGA_3730 coding for a hemolysin-type calcium-binding repeat containing protein, a homolog of which is found in Xylella strains with 55% sequence identity. In Xylella, this gene is annotated as a member of a family of pore forming toxins/RTX toxins. Its homolog is also found in other plant pathogens (i.e. P. syringae pv. syringae B728a and R. solanacearum GMI1000). This protein has been described as a type I effector in X. fastidiosa strain temecula (PD1506) [92]. RTX toxin family members, especially of the hemolysin type, have been shown to be virulence factors in a variety of cell types in eukaryotes [93, 94]. Finally, a gene XGA_0603 coding for lanthionine synthetase (lantibiotic biosynthesis) is found among these Xg specific genes, a homolog of which is found in Xvm NCPPB702. LanL enzymes in pathogenic bacteria contribute to virulence by modifying the host signaling pathways, in most cases by inactivating MAPKs [95].
Genes common to all tomato pathogens but absent from other sequenced xanthomonads
In order to see what defines the tomato pathogens, we compared the four sequenced genomes (Xv, Xp, Xg and Xcv) to other sequenced xanthomonads. We found seven genes that were conserved in all four tomato pathogens and absent from most of other sequenced xanthomonads with the exception of Xcm, Xvv, Xaub and Xauc, which possess homologs for six out of these seven genes (Table 8). Only the hypothetical protein XCV2641 seems to be specific to the four tomato pathogens. This gene shows only 35% sequence identity to a gene from Xvv and Xcm. A homolog of the hypothetical protein, XCV4416 was found in Xau, but is absent from all other sequenced xanthomonads. Genes homologous in Xcm and Xvv include two transposase genes both belonging to the transposase 17 superfamily (XCV0615, XCV0623), XCV0041 (putative penicillin amidase fragment), XCV0111 (lignostilbene-alpha, beta dioxygenase), XCV0112 (uncharacterized protein conserved in bacteria) (Table 8). Interestingly, XCV0111 encodes a protein known to be involved in phenylpropanoid degradation. Phenylpropanoids are well known plant secondary metabolites induced during defense response upon pathogen attack [96]. It appears that the four tomato pathogens along with Xvv and Xcm have acquired this function to disarm the basal plant defense.
The evolution of pathogenicity clusters corresponds to the MLST-based phylogeny
The correlation between tree topology using MLST and phylogeny based on the sequences of pathogenicity clusters and the avrBs2 effector gene, which is found in all xanthomonads, was tested. Based on MLST, Xp and Xcv group together along with Xac while Xg is more closely related to Xcc. Xv forms a different clade and is more closely related to the Xcc group. As can be seen in Figure 7, phylogeny based on MLST is congruent with phylogeny based on the pathogenicity clusters (gum, hrp cluster) and based on the avrBs2 effector, suggesting that overall these clusters were vertically inherited from the most recent common ancestor of these strains.
Correlation between phylogenies based on Multi-Locus Sequence Typing (MLST) core genome and pathogenicity clusters: Concatenated amino acid sequences of the six genes fusA, gapA, gltA, gyrB, lacF, lepA from four bacterial spot pathogen strains along with other sequenced xanthomonads are considered in the analysis. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. Phylogenetic analyses were conducted in MEGA4.
Conclusions
The interaction of Xanthomonas strains with tomato and pepper represents a model system for studying plant-pathogen co-evolution because of the diversity present among the strains causing bacterial spot. Although the four Xanthomonas species infect the same host, tomato, and cause very similar disease, they are genetically diverse pathogens. The comparative genomic analysis has provided insights into the evolution of these strains. Whole genome comparisons revealed that Xg and Xv are more closely related to Xcc than Xcv and Xp. A few pathogenicity clusters, such as hrp, xcs and xps of Xg and Xv, were similar in terms of genetic organization and sequence identity to Xcc (Figure 8). However, a few pathogenicity clusters of the four strains belonging to four phylogenetic groups showed different evolutionary origins. While the pepper pathogens Xcv, Xv and Xg possess similar LPS biosynthesis cluster, part of the LPS cluster from Xp is similar to the one from Xac (Figure 8). Xv contains few effectors, including xopAG (avrGf1) and xopAI the latter of which was previously found to be unique to citrus pathogens Xac, Xaub and Xauc[25]. Xg has a number of effectors homologous to P. syringae type III effectors suggesting probable horizontal transfer of these effectors. Xg contains a unique T4SS along with the one that is exclusive to Xp, Xv and Xg. Xp has two T6SSs, as found in Xcv. Xv has only one T6SS which is similar to that of Xac. Xg has no T6SS as seen for Xcc (Figure 8). While Xg and Xv show close relationship to Xcc based on whole genome comparisons, few pathogenicity clusters mentioned above seem to be conserved among tomato/pepper xanthomonads.
A diagrammatic representation of relationship among bacterial spot xanthomonads, Xac and Xcc with respect to presence or absence of pathogenicity clusters. Similar color shade indicates high identity and similar cluster organization. Lower sequence identities compared to the reference are indicated by faded gray shades. Reference strain is indicated by asterisk next to the symbol. The absence of certain part of cluster is indicated by white. In the case of LPS cluster, Xv and Xac contain novel cluster regions in the C terminal region which is indicated by a different color. Xac and Xcv contain a plasmid borne type IV cluster. Although it differs from type IVA present in other bacterial spot xanthomonads, Xac and Xcv cluster is mentioned here under type IVA with different colors. A blank space indicates complete absence of gene cluster in that particular species. A more detailed representation of individual clusters can be found in figures 2 through 5.
Type III effectors have been investigated for their contribution to pathogenicity and host-range specificity. In addition to homologs of the known effectors, we identified novel effectors in the draft genomes. By comparing effector repertoires of tomato pathogens, two possible candidate pathogenicity determinants, xopF1 and xopD, were identified, of which xopD is responsible for delaying symptom development, and in turn, is important for pathogen survival. Unique genes present in Xg include the novel effectors xopAO, xopAQ, xopAS and an avrBs1 member as well as a few other virulence factors, which have been characterized in other plant pathogens and which could explain the aggressive nature of Xg on pepper. Each species contains at least three unique type III effectors, which could explain host preferences among the strains and their aggressiveness on tomato/pepper. Comparison of the LPS clusters between the four species revealed significant variation. Xp has acquired a novel LPS cluster during evolution, which might be responsible for its predominance and its limited host range. As seen from the in planta growth assay of Xp ΔavrXv3 mutant carrying the LPS O-antigen from Xcv, the LPS cluster from pepper pathogens can be a contributor to the increased in planta growth of Xp ΔavrXv3 mutant on pepper, but is not the absolute virulence determinant. Use of the XA21 receptor similar to the Xoo-rice system in Xcv - tomato/pepper could be one of the ways to confer resistance to xanthomonads due to presence of a similar AX21 peptide and a functional rax system in Xcv. Common and unique genes encoding enzymes involved in cell wall deconstruction are candidates for further study to define host preference and virulence.
In conclusion, comparison of draft genomes obtained by next generation sequencing has allowed an in-depth study of diverse groups of bacterial spot pathogens at the genomic level. This analysis will serve as a basis to infer evolution of new virulent strains and overcoming existing host resistance. The knowledge of potential virulence or pathogenicity factors is expected to aid in devising effective control strategies and breeding for durable resistance in tomato and pepper cultivars.
Methods
Genome sequencing
Xv, Xp and Xg were sequenced by 454-pyrosequencing [27] at core DNA sequencing facility, ICBR, University of Florida. Xanthomonas isolates were grown overnight in nutrient broth. Genomic DNA was isolated using CTAB-NaCl extraction method [97] and resuspended in TE buffer (10 mM Tris pH 8, 1 mM EDTA pH 8). Libraries of fragmented genomics DNA were sequenced on 454-Genome Sequencer, FLX instrument at Interdisciplinary Center for Biotechnology Research (ICBR) at UF. De novo assemblies were constructed using 454 Newbler Assembler [27]. The three draft genomes were obtained with around 10× coverage.
For Illumina sequencing, the Xanthomonas strains were purified from single-colony and grown overnight in liquid cultures. Genomic DNA was isolated by phenol extraction and precipitated twice with isopropanol, and finally dissolved in TE buffer. DNA was then purified by cesium chloride density gradient centrifugation and precipitated with 95% ethanol, then dissolved in TE buffer. Libraries of fragmented genomic DNA with adapters for paired-end sequencing were prepared according to the protocol provided by Illumina, Inc. with minor modifications. The libraries were sequenced on the 2G Genome Analyzer at Center of Genome Research & Biocomputing at Oregon State University and post-processed using a standard Illumina pipeline [28]. We obtained approximately 8-10 million 60-bp reads for each genome, providing roughly 95× predicted coverage.
Assembly and annotation
De novo assembly was generated on Newbler assembler (version 2.3; 454 Life Science, Branford, CT) using 454-sequencing reads for each genome. CLC workbench [29] was used in the next step for combining 454-based contigs with illumina reads, wherein, 454 based contigs were used as long reads to fill in gaps generated during combined de novo assembly. These combined assemblies of each genome were uploaded on IMG-JGI (Joint Genome Institute, Walnut Creek, California) server for gene calling. The gene prediction was carried out using GeneMark. Pfam, InterPro, COGs assignments were carried out for identified genes. Pathogenicity clusters described in the paper were manually annotated.
Whole genome comparisons
We aligned draft genomes against reference Xanthomonas genomes using nucmer [31] of MUMmer program (version 3.20) and dnadiff was used to calculate percentage of aligned sequences. We have also compared genomes using the MUM index [30] to measure distances between two genomes. The maximal unique exact matches index (MUMi) distance calculation was performed using the Mummer program (version 3.20). Mummer was run on concatenated contigs or replicons (achieved by inserting a string of 20 symbols 'N' between contig or replicon sequences) of each genome. The distance calculations performed using the MUMi script are based on the number of maximal unique matches of a given minimal length shared by two genomes being compared. MUMi values vary from 0 for identical genomes to 1 for very distant genomes [30].
Phylogenetic analysis
MLST sequences (fusA, gapA, gltA, gyrB, lacF, lepA) for all the genomes were obtained in concatenated form from PAMDB website http://pamdb.org. Genes and their corresponding amino acid sequences spanning gum, hrp cluster were downloaded from NCBI genbank sequences of sequenced genomes. Amino acid sequences of proteins of these clusters for Xcv and Xcc were used as query to search for homology against draft genomes of Xp, Xv and Xg. The amino acid sequences were then concatenated for each pathogenicity cluster and then aligned using CLUSTALW ignoring gaps. Neighbour-joining trees were constructed with boostrap value for 1000 replicates using MEGA4 [98]. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 2723 positions in the final dataset.
Phylogeny reconstruction
Species tree. We used a supermatrix approach as in previous work [25]. Protein sequences of six Xanthomonas genomes (ingroups) and the S. maltophilia R551-3 genome (outgroup) were clustered in 5,096 families using OrthoMCL [99]. We then selected families with one and only one representative from each of the ingroup genomes and at most one outgroup protein, resulting in 2,282 families. Their sequences were aligned using MUSCLE [100] and the resulting alignments were concatenated. Non-informative columns were removed using Gblocks [101], resulting in 792,079 positions. RAxML [102] with the PROTGAMMAWAGF model was used to build the final tree.
Prediction of effector repertoires, cloning of candidate effectors and confirmation using avrBs2 reporter gene assay
A database was created collecting all the known plant and animal pathogen effectors. Using all these known effectors as query, tblastn analysis was performed against all contigs of the draft genomes of Xv, Xg and Xp with e-value of 10-5[103]. Pfam domains were searched for possible domains found in known effectors in predicted set of ORFs of draft genome sequences. Candidate effectors were classified according to the nomenclature and classification scheme for effectors in xanthomonads recently [24]. Candidate effectors showing < 45% identity at amino acid level to the known effectors were confirmed for their translocation using avrBs2 reporter gene assay.
N-terminal 100 amino acid region along with upstream 500 bps sequence of candidate genes were PCR amplified using primers with BglII restriction sites at the 5' ends. Following digestion with BglII, PCR amplicons were ligated with BglII-digested pBS(BglII::avrBs262-574::HA) (courtesy of Dr. Mary Beth Mudgett, Stanford university), and later transformed into E. coli DH5α. In-frame fusions were confirmed by DNA sequencing using F20 and R24 primers. BamHI-KpnI fragments containing the candidate gene fused to avrBs2 was then cloned into pUFR034. Resulting plasmids were then introduced into Xcv pepper race 6 (TED3 containing mutation in avrBs2) by tri-parental mating. The resulting Xcv strains were inoculated on Bs2 pepper cv. ECW 20R and kept at 28°C in growth room. After 24 hours, strong HR was indicating successful translocation of candidate effector fusions.
Cloning of pepper specificity genes in Xp
The three genes mentioned above were cloned individually and in combination in pLAFR3 vector and conjugated in Xp 91-118 ΔavrXv3 mutant PM1. The PM1 transconjugants with the three individual genes and combined ones along with virulent pepper race 6 strain were infiltrated at 105 CFU/ml concentration in pepper cv. ECW and leaves were sampled at every 48 hours after inoculation. The samples were plated on nutrient agar, incubated at 27°C and CFU/ml counts were enumerated. Experiment was carried out in triplicate and repeated three times.
Database submission
The draft genome sequences of Xanthomonas vesicatoria ATCC 35937 (Xv) have been deposited at DDBJ/EMBL/GenBank under accession number AEQV00000000. The draft genome sequences of Xanthomonas perforans 91-118 (Xp) have been deposited at DDBJ/EMBL/GenBank under accession number AEQW00000000. The draft genome sequences of Xanthomonas gardneri ATCC 19865 (Xg) have been deposited at DDBJ/EMBL/GenBank under accession number AEQX00000000. The version described in this paper is the first version, AEQV01000000, AEQW01000000, AEQX01000000. All three draft genomes will be released upon manuscript acceptance.
Abbreviations
- Xcv Xanthomonas euvesicatoria :
-
strain 85-10
- formerly:
-
Xanthomonas campestris pv. vesicatoria strain 85-10
- Xv Xanthomonas vesicatoria :
-
strain 1111 (ATCC 35937)
- Xp Xanthomonas perforans :
-
strain 91-118
- Xg Xanthomonas gardneri :
-
strain 101 (ATCC 19865)
- Xvv Xanthomonas vasicola :
-
pv. vasculorum NCPPB702
- Xac Xanthomonas citri :
-
subsp. citri strain 306
- formerly:
-
Xanthomonas axonopodis pv. citri strain 306
- Xaub Xanthomonas fuscans :
-
subsp. aurantifolii B strain
- Xauc Xanthomonas fuscans :
-
subsp. aurantifolii C strain
- Xalb:
-
Xanthomonas albilineans
References
Pohronezny K, Volin RB: The effect of bacterial spot on yield and quality of fresh market tomatoes. Hort Science. 1983, 18: 69-70.
Jones JB, Stall RE, Bouzar H: Diversity among xanthomonads pathogenic on pepper and tomato. Annu Rev Phytopathol. 1998, 36: 41-58. 10.1146/annurev.phyto.36.1.41.
Jones JB, Bouzar H, Stall RE, Almira EC, Roberts PD, Bowen BW, Sudberry J, Strickler PM, Chun J: Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int J Syst Evol Microbiol. 2000, 50: 1211-1219.
Bouzar H, Jones JB, Somodi GC, Stall RE, Daouzli N, Lambe RC, Felix Gastelum R, Trinidad Correa R: Diversity of Xanthomonas campestris pv. vesicatoria in tomato and pepper fields of Mexico. Can J Plant Pathol. 1996, 18: 75-77. 10.1080/07060669609500659.
Bouzar H, Jones JB, Stall RE, Louws FJ, Schneider M, Rademaker JLW, de Bruijn FJ, Jackson LE: Multiphasic analysis of xanthomonads causing bacterial spot disease on tomato and pepper in the Caribbean and Central America: Evidence for common lineages within and between countries. Phytopathology. 1999, 89: 328-335. 10.1094/PHYTO.1999.89.4.328.
Kim SH, Olson TN, Peffer ND, Nikolaeva EV, Park S, Kang S: First report of bacterial spot of tomato caused by Xanthomonas gardneri in Pennsylvania. Plant Disease. 2010, 94: 638-10.1094/PDIS-94-5-0638B.
Hamza AA, Robene-Soustrade I, Jouen E, Gagnevin L, Lefeuvre P, Chiroleu F, Pruvost O: Genetic and pathological diversity among Xanthomonas strains responsible for bacterial spot on tomato and pepper in the southwest Indian Ocean region. Plant Disease. 2010, 94: 993-999. 10.1094/PDIS-94-8-0993.
Myung IS, Moon SY, Jeong IH, Lee YK, Lee YH, Ra DS: Bacterial spot of tomato caused by Xanthomonas perforans, a new disease in Korea. Plant Disease. 2009, 93: 1349-10.1094/PDIS-93-12-1349B.
Sutic D: Bakterioze crvenog patlidzana (Tomato bacteriosis). Posebna Izd Inst Zasht Bilja Beograd. 1957, 6: 1-65. (special edition). Beograd: Institute of Plant Protein. (English summary Rev Appl Mycol 1957, 36: 734-735.)
De Ley J: Modern molecular methods in bacterial taxonomy: evaluation, application, prospects. Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, Angers. 1978, 347-357.
Jones JB, Lacy GH, Bouzar H, Stall RE, Schaad NW: Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol. 2004, 27: 755-762. 10.1078/0723202042369884.
Almeida NF, Yan S, Cai R, Clarke CR, Morris CE, Schaad NW, Schuenzel EL, Lacy GH, Sun X, Jones JB, Castillo JA, Bull CT, Leman S, Guttman DS, Setubal JC, Vinatzer BA: PAMDB, A multilocus sequence typing and analysis database and website for plant-associated microbes. Phytopathology. 2010, 100: 208-215. 10.1094/PHYTO-100-3-0208.
Obradovic A, Jones JB, Momol MT, Balogh B, Olson SM: Management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Disease. 2004, 88: 736-740. 10.1094/PDIS.2004.88.7.736.
Louws FJ, Wilson M, Campbell HL, Cuppels DA, Jones JB, Shoemaker PB, Sahin F, Miller SA: Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Disease. 2001, 85: 481-488. 10.1094/PDIS.2001.85.5.481.
Kearney B, Staskawicz BJ: Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature. 1990, 346: 385-386. 10.1038/346385a0.
Gassmann W, Dahlbeck D, Cjesnokova O, Minsavage GV, Jones JB, Staskawicz BJ: Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria. J Bacteriol. 2000, 182: 7053-7059. 10.1128/JB.182.24.7053-7059.2000.
Stall RE, Jones JB, Minsavage GV: Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu Rev Phytopathol. 2009, 47: 265-84. 10.1146/annurev-phyto-080508-081752.
Bonas U, Schulte R, Fenselau S, Minsavage GV, Staskawicz BJ, Stall RE: Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and hypersensitive response on pepper and tomato. Mol Plant Microbe Interact. 1991, 4: 81-88. 10.1094/MPMI-4-081.
Kim JG, Park BK, Yoo CH, Jeon E, Oh J, Hwang I: Characterization of the Xanthomonas axonopodis pv. glycines Hrp Pathogenicity Island. J Bacteriol. 2003, 185: 3155-3166. 10.1128/JB.185.10.3155-3166.2003.
Nimura K, Melotto M, He S: Suppression of host defense in compatible plant- Pseudomonas syringae interactions. Curr Opinion Plant Biol. 2005, 8: 361-368. 10.1016/j.pbi.2005.05.005.
Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL: Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006, 60: 425-449. 10.1146/annurev.micro.60.080805.142251.
Sarkar S, Gordon J, Martin G, Guttman D: Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics. 2006, 174: 1041-1056. 10.1534/genetics.106.060996.
Rohmer L, Guttman DS, Dangl JL: Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae. Genetics. 2004, 167: 1341-1360. 10.1534/genetics.103.019638.
White FF, Potnis N, Jones JB, Koebnik R: The Type III effectors of Xanthomonas. Mol Plant Pathol. 2009, 10: 749-766. 10.1111/j.1364-3703.2009.00590.x.
Moreira LM, Almeida NF, Potnis N, Digiampietri LA, Adi SS, Bortolossi JC, da Silva AC, da Silva AM, de Moraes FE, de Oliveira JC, de Souza RF, Facincani AP, Ferraz AL, Ferro MI, Furlan LR, Gimenez DF, Jones JB, Kitajima EW, Laia ML, Leite RP, Nishiyama MY, Neto JR, Nociti LA, Norman DJ, Ostroski EH, Pereira HA, Staskawicz BJ, Tezza RI, Ferro JA, Vinatzer BA, Setubal JC: Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics. 2010, 11: 238-10.1186/1471-2164-11-238.
Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Büttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, Kirchner O, Lanz C, Linke B, McHardy AC, Meyer F, Mittenhuber G, Nies DH, Niesbach-Klösgen U, Patschkowski T, Rückert C, Rupp O, Schneiker S, Schuster SC, Vorhölter FJ, Weber E, Pühler A, Bonas U, Bartels D, Kaiser O: Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol. 2005, 187: 7254-7266. 10.1128/JB.187.21.7254-7266.2005.
Margulies M, Egholm M, Altman W, Attiya S, Bader J, Bemden L, Berka J, Braverman M, Chen Y, Chen Z, Dewell S, Du L, Fierro J, Gomes X, Begley R, Rothberg J: Genome sequencing in open microfabricated high density picoliter reactions. Nature. 2005, 437: 376-380.
Bentley DR: Whole genome resequencing. Curr Opinion Genet Dev. 2006, 16: 545-552. 10.1016/j.gde.2006.10.009.
CLC Genomics Workbench: White paper on de novo assembly in CLC NGS Cell 3.0 beta. 2010, [http://www.clcbio.com]
Delonger M, Karoui ME, Petit M: A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol. 2009, 191: 91-99. 10.1128/JB.01202-08.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL: Versatile and open software for comparing large genomes. Genome Biol. 2004, 5: R12-10.1186/gb-2004-5-2-r12.
Kim JG, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, McLane H, Martin GB, Mudgett MB: Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell. 2009, 21: 1305-1323. 10.1105/tpc.108.063123.
Büttner D, Noel L, Stutmann J, Bonas U: Characterization of the nonconserved hpaB-hrpF region in the hrp pathogenicity island from Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact. 2007, 20: 1063-1074.
Silva ACR, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio R, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LMC, Amaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EG, Lemos MV, Locali EC, Machado MA, Madeira AM, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CF, Miyaki CY, Moon DH, Moreira LM, Novo MT, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, Rossi A, Sena JA, Silva C, de Souza RF, Spinola LA, Takita MA, Tamura RE, Teixeira EC, Tezza RI, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP: Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002, 417: 459-463. 10.1038/417459a.
Stavrinides J, Ma W, Guttman D: Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathogens. 2006, 2: e104-10.1371/journal.ppat.0020104.
Kim JG, Taylor KW, Hotson A, Keegan M, Schmelz EA, Mudgett MB: XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell. 2008, 20: 1915-1929. 10.1105/tpc.108.058529.
Astua-Monge G, Minsavage GV, Stall RE, Davis MJ, Bonas U, Jones JB: Resistance of tomato and pepper to T3 strains of Xanthomona campestris pv. vesicatoria is specified by a plant-inducible avirulence gene. Mol Plant Microbe Interact. 2000, 13: 911-921. 10.1094/MPMI.2000.13.9.911.
Astua-Monge G, Minsavage GV, Stall RE, Vallejos CE, Davis MJ, Jones JB: Xv4-Avrxv4: A new gene-for-gene interaction identified between Xanthomonas campetris pv. vesicatoria race T3 and the wild tomato relative Lycopersicon pennellii. Mol Plant Microbe Interact. 2000, 13: 1346-1355. 10.1094/MPMI.2000.13.12.1346.
Rybak M, Minsavage GV, Stall RE, Jones JB: Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol Plant Pathol. 2009, 10: 249-262. 10.1111/j.1364-3703.2008.00528.x.
Minsavage GV, Dahlbeck D, Whalen MC, Kearny B, Bonas U, Staskawicz BJ, Stall RE: Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria- pepper interactions. Mol Plant Microbe Interact. 1990, 3: 41-47. 10.1094/MPMI-3-041.
Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T: Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 2008, 179: 546-556. 10.1111/j.1469-8137.2008.02487.x.
Noel L, Thieme F, Nennstiel D, Bonas U: C-DNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol Microbiol. 2001, 41: 1271-1281. 10.1046/j.1365-2958.2001.02567.x.
Araujo ER, Pereira RC, Moita AW, Ferreira MASV, Café-Fiho AC, Quezado-Duval AM: Effect of temperature on pathogenicity components of tomato bacterial spot and competition between Xanthomonas perforans and X. garnderi. III International symposium on tomato diseases. 2010
Vernikos GS, Parkhill J: Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006, 22: 2196-203. 10.1093/bioinformatics/btl369.
Lorang JM, Shen H, Kobayashi D, Cooksey D, Keen NT: AvrA and avrE in Pseudomonas syrinage pv. tomato PT23 play a role in virulence on tomato plants. Mol Plant Microbe Interact. 1994, 7: 508-515. 10.1094/MPMI-7-0508.
Mukaihara T, Tamura N, Iwabuchi M: Genome-wide identification of large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol Plant Microbe Interact. 2010, 23: 251-262. 10.1094/MPMI-23-3-0251.
Lee S.-W, Jeong K.-S, Han S.-W, Lee S.-E, Phee B.-K, Hahn T.-R, Ronald P: The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. J Bacteriol. 2008, 190: 2183-2197. 10.1128/JB.01406-07.
Lee S.-W, Han S.-W, Sririyanum M, Park C.-J, Seo Y.-S, Ronald PC: A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 2009, 326: 850-853. 10.1126/science.1173438.
da Silva FG, Shen Y, Dardick C, Burdman S, Yadav RC, de Leon AL, Ronald PC: Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol Plant Microbe Interact. 2004, 17: 593-601. 10.1094/MPMI.2004.17.6.593.
Jha G, Rajeshwari R, Sonti R: Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol Plant Microbe Interact. 2005, 18: 891-898. 10.1094/MPMI-18-0891.
Wang L, Rong W, He C: Two Xanthomonas extracellular polygalacturonases, PghAxc and PghBxc, are regulated by type III secretion regulators HrpX and HrpG and are required for virulence. Mol Plant Microbe Interact. 2008, 21: 555-563. 10.1094/MPMI-21-5-0555.
Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, Bonas U, Büttner D: Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria. New Phytol. 2010, 187: 983-1002. 10.1111/j.1469-8137.2010.03312.x.
Hurlbert JC, Preston JF: Functional characterization of a novel xylanase from corn strains of Erwinia chrysanthemi. J Bacteriol. 2001, 183: 2093-2100. 10.1128/JB.183.6.2093-2100.2001.
St John FJ, Rice JD, Preston JF: Characterization of XynC from Bacillus subtilis subspecies subtilis strain 168 and Analysis of Its Role in Depolymerization of Glucuronoxylan. J Bacteriol. 2006, 24: 8617-8626. 10.1128/JB.01283-06.
Sun Q, Hu J, Huang G, Ge C, Fang R, He C: Type-II secretion pathway structural gene xpsE, xylanase- and cellulase secretion and virulence in Xanthomonas oryzae pv. oryzae. Plant Pathol. 2005, 54: 15-21. 10.1111/j.1365-3059.2004.01101.x.
Goesaert H, Gebruers K, Brijs K, Courtin CM, Delcour JA: XIP-type endoxylanase inhibitors in different cereals. J Cereal Sci. 2003, 38: 317-324. 10.1016/S0733-5210(03)00046-8.
Rajeshwari R, Jha G, Sonti R: Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol Plant Microbe Interact. 2005, 18: 830-837. 10.1094/MPMI-18-0830.
Biely P, Vrsanska MM, Tenkanen M, Kluepfel D: Endo-beta-1,4-xylanase families: differences in catalytic properties. J Biotechnol. 1997, 57: 151-166. 10.1016/S0168-1656(97)00096-5.
Preston JF, Hurlbert JC, Rice JD, Ragunathan A, St John FJ: Microbial Strategies for the Depolymerization of Glucuronoxylan: Leads to the Biotechnological Applications of Endoxylanases. Application of Enzymes to Lignocellulosics. Edited by: Mansfield SD, Saddler JN. 2003, ACS Symposium Series No. 855, Ch 12: 191-210. full_text.
Shulami S, Gal O, Sonenshein AL, Shoham Y: The glucuronic acid-utilization gene cluster from Bacillus stearothermophilus T-6. J Bacteriol. 1999, 181: 3695-3704.
Shulami S, Zaide G, Zolotnitsky G, Langut Y, Feld G, Sonenshein AL, Shoham Y: A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl Environ Microbiol. 2007, 73: 874-84. 10.1128/AEM.02367-06.
Chow V, Nong G, Preston JF: Structure, function and regulation of the aldouronate-utilization gene cluster from Paenibacillus sp. JDR-2. J Bacteriol. 2007, 189: 8863-8870. 10.1128/JB.01141-07.
El Tahir Y, Kuusela P, Skurnik M: Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG -S motifs on the amino-terminal half of the protein involved in collagen binding. Mol Microbiol. 2000, 37: 192-206. 10.1046/j.1365-2958.2000.01992.x.
Das A, Rangaraj N, Sonti R: Multiple adhesion-like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry and virulence on rice. Mol Plant Microbe Interact. 2009, 22: 73-85. 10.1094/MPMI-22-1-0073.
Wang X, Preston JF, Romeo T: The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004, 186: 2724-2734. 10.1128/JB.186.9.2724-2734.2004.
Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I: Dissecting the bacterial type VI secretion system by genome wide in silico analysis: what can be learned from available microbial genomic resources?. BMC Genomics. 2009, 10: 104-10.1186/1471-2164-10-104.
Vorhölter FJ, Niehaus K, Pühler A: Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol Genet Genomics. 2001, 266: 79-95.
Patil PB, Bogdanove AJ, Sonti RV: The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol Biol. 2007, 7: 243-10.1186/1471-2148-7-243.
Molinaro A, Silipo A, Lanzetta R, Newmann M, Dow M, Parrilli M: Structural elucidation of the O-chain of the lipopolysaccharide from Xanthomonas campestris strain 8004. Carbohydr Res. 2003, 338: 277-281. 10.1016/S0008-6215(02)00433-0.
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A: The Pfam protein families database. Nucleic Acid Research Database issue. 2010, 38: D211-222. 10.1093/nar/gkp985.
Jones JB, Bouzar H, Somodi GC, Stall RE, Pernezny K, El-Morsy G, Scott JW: Evidence for the Preemptive Nature of Tomato Race 3 of Xanthomonas campestris pv. vesicatoria in Florida. Phytopathology. 1998, 88: 33-38. 10.1094/PHYTO.1998.88.1.33.
Mooi FR, Bik EM: The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 1997, 5: 161-165. 10.1016/S0966-842X(96)10086-X.
Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM: A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol. 2000, 38: 986-1003. 10.1046/j.1365-2958.2000.02196.x.
He YW, Zhang LH: Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol Rev. 2008, 32: 842-857. 10.1111/j.1574-6976.2008.00120.x.
Dow M: Diversification of the function of cell-to-cell signaling in regulation of virulence within plant pathogenic xanthomonads. Sci Signal. 2008, 1: 23-10.1126/stke.121pe23.
Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM: Cell-cell signal dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc Natl Acad Sci USA. 2010, 107: 5989-5994. 10.1073/pnas.0912839107.
Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJG, Slater H, Dow JM, Williams P, and Daniels MJ: A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997, 24: 555-566. 10.1046/j.1365-2958.1997.3721736.x.
Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL: Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci USA. 2003, 100: 10995-11000. 10.1073/pnas.1833360100.
Chatterjee S, Sonti RV: rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol Plant Microbe Interact. 2002, 15: 463-471. 10.1094/MPMI.2002.15.5.463.
Siciliano F, Torres P, Sendín L, Bermejo C, Filippone P, Vellice G, Ramallo J, Castagnaro A: Analysis of the molecular basis of Xanthomonas axonopodis pv. citri pathogenesis in Citrus limon. Electron J Biotechnol. 2006, 9: 10.2225/vol9-issue3-fulltext-20.
Wang L, Makino S, Subedee A, Bogdanove AJ: Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis. Appl Environ Microbiol. 2007, 73: 8023-8027. 10.1128/AEM.01414-07.
Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM: Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA. 2006, 103: 6712-6717. 10.1073/pnas.0600345103.
Romling U, Gomelsky M, Galperin MY: C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005, 57: 629-639. 10.1111/j.1365-2958.2005.04697.x.
Jenal U, Malone J: Mechanisms of Cyclic-di-GMP Signaling in Bacteria. Annu Rev Genet. 2006, 40: 385-407. 10.1146/annurev.genet.40.110405.090423.
Hengge R: Principles of c-di-GMP signalling in bacteria. Nature Rev Microbiol. 2009, 7: 263-273. 10.1038/nrmicro2109.
Ryan RP, Fouhy Y, Lucey JF, Jiang BL, He YQ, Feng JX, Tang JL, Dow JM: Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol Microbiol. 2007, 63: 429-442. 10.1111/j.1365-2958.2006.05531.x.
He YW, Boon C, Zhou L, Zhang LH: Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol Microbiol. 2009, 71: 1464-1476. 10.1111/j.1365-2958.2009.06617.x.
Crossman VC, Gould JM, Dow GS, Vernikos A, Okazaki M, Sebaihia D, Saunders C, Arrowsmith T, Carver N, Peters E, Adlem A, Kerhornou A, Lord L, Murphy K, Seeger R, Squares S, Rutter MA, Quail MA, Rajandream D, Harris C, Churcher SD, Bentley J, Parkhill NR, Avison MB: The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008, 9: R74-10.1186/gb-2008-9-4-r74.
Hert AP, Roberts PD, Momol MT, Minsavage GV, Tudor-Nelson SM, Jones JB: Relative importance of bacteriocin-like genes in antagonism of Xanthomonas perforans tomato race 3 to Xanthomonas euvesicatoria tomato race 1 strains. Appl Environ Microbiol. 2005, 71: 3581-3588. 10.1128/AEM.71.7.3581-3588.2005.
Tudor-Nelson SM, Minsavage GV, Stall RE, Jones JB: Bacteriocin-like substances from tomato race 3 strains of Xanthomonas campestris pv. vesicatoria. Phytopathology. 2003, 93: 1415-1421. 10.1094/PHYTO.2003.93.11.1415.
Alavi SM, Sanjari S, Durand F, Brin C, Manceau C, Poussier S: Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl Environ Microbiol. 2008, 74: 3295-3301. 10.1128/AEM.02507-07.
Reddy JD, Reddy SL, Hopkins DL, Gabriel DW: TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol Plant Microbe Interact. 2007, 20: 403-410. 10.1094/MPMI-20-4-0403.
Lally ET, Hill RB, Kieba LR, Korstoff J: The interaction between RTX toxins and target cells. Trends Microbiol. 1999, 7: 356-361. 10.1016/S0966-842X(99)01530-9.
Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkin I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P: RTX-toxins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010, 34: 1076-1112.
Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA: Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010, 8: e1000339-10.1371/journal.pbio.1000339.
Dixon RA, Achnine L, Kota P, Liu C, Reddy MS, Wang L: The phenylpropanoid pathway and plant defence - a genomics perspective. Mol Plant Pathol. 2002, 3: 371-390. 10.1046/j.1364-3703.2002.00131.x.
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K: Current protocols in molecular biology. 1994, New York, N. Y.: John Wiley and Sons, 1: 2.4.1-2.4.2.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599. 10.1093/molbev/msm092.
Li L, Stoeckert CJ, Roos DS: OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13: 2178-2189. 10.1101/gr.1224503.
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32: 1792-1797. 10.1093/nar/gkh340.
Castresana J: Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, 17: 540-552.
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Altschul SF, Madeen TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Kearney B, Staskawicz BJ: Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature. 1990, 346: 385-386. 10.1038/346385a0.
Roden JA, Belt B, Ross JB, Tachibana T, Vargas J, Mudgett MB: A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc Natl Acad Sci USA. 2004, 101: 16624-16629. 10.1073/pnas.0407383101.
Furutani A, Takaoka M, Sanada H, Noguchi Y, Oku T, Tsuno K, Ochiai H, Tsuge S: Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 2009, 22: 96-106. 10.1094/MPMI-22-1-0096.
Jiang W, Jiang BL, Xu RQ, Huang JD, Wei HY, Jiang GF, Cen WJ, Liu J, Ge YY, Li GH, Su LL, Hang XH Tang DJ, Lu GT, Feng JX, He YQ, Tang JL: Identification of six type III effector genes with PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol Plant Microbe Interact. 2009, 22: 1401-1411. 10.1094/MPMI-22-11-1401.
Kim JG, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, McLane H, Martin GB, Mudgett MB: Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell. 2009, 21: 1305-1323. 10.1105/tpc.108.063123.
Metz M, Dahlbeck D, Morales CQ, Al Sady B, Clark ET, Staskawicz BJ: The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 2005, 41: 801-814. 10.1111/j.1365-313X.2005.02338.x.
Guidot A, Prior P, Schoenfeld J, Carrere S, Genin S, Boucher C: Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum from gene distribution analysis. J Bacteriol. 2007, 189: 377-387. 10.1128/JB.00999-06.
Park DS, Hyun JW, Park YJ, Kim JS, Kang HW, Hahn JH, Go SJ: Sensitive and specific detection of Xanthomonas axonopodis pv. citri by PCR using pathovar specific primers based on HrpW gene sequences. Microbiol Res. 2006, 161: 145-149. 10.1016/j.micres.2005.07.005.
Xu RQ, Li XZ, Wei HY, Jiang B, Li K, He YQ, Feng JX, Tang JL: Regulation of eight avr genes by hrpG and hrpX in Xanthomonas campestris pv. campestris and their role in pathogenicity. Progress in Natural Science. 2006, 16: 1288-1294. 10.1080/10020070612330014.
Noel L, Thieme F, Nennstiel D, Bonas U: C-DNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol Microbiol. 2001, 41: 1271-1281. 10.1046/j.1365-2958.2001.02567.x.
Thieme F, Szczesny R, Urban A, Kirchner O, Hause G, Bonas U: New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol Plant Microbe Interact. 2007, 20: 1250-1261. 10.1094/MPMI-20-10-1250.
Thieme F: Genombasierte Identifizierung neuer potentieller Virulenzfaktoren von Xanthomonas campestris pv. vesicatoria. Thesis. 2008, Mathematisch-Naturwissenschaftlich-Technische Fakultät der Martin-Luther Universität, Halle-Wittenberg
Acknowledgements
This research was supported by USDA-NIFA special grant T-STAR (J. B. Jones, J. F. Preston and M. T. Momol, USDA 2006-34135-17569). We thank IMG-JGI for providing genome annotation pipeline and interface for the comparative genomic analyses. Special thanks to Kostantinos Mavromatis in helping preparation of Genbank files.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
JBJ conceived the project. JBJ and BJS oversaw genomic sequencing. JBJ provided the strains. KK did genome assembly. NP, RPR, RK, VC, PBP, FB annotated pathogenicity clusters in the genomes. DJN and BJS helped create an effector database. NP carried out effector analysis and confirmed them experimentally. NP, BAV, RK, FFW and JBJ interpreted effector analysis. NP, RPR, RK, VC, JFP, PBP, JMD, MS, TM did analyses of different pathogenicity clusters of the three genomes and helped writing corresponding sections in the manuscript. NP oversaw the experimental validations of the pathogenicity clusters. NFA created ortholog families, and did phylogenetic analysis based on all orthologus families of the three draft genomes and the reference genomes. JCS did MUMi analysis and constructed phylogenetic tree based on that analysis. NP generated the GenBank files. DJN, BJS, FFW, RK, JBJ, BAV, JFP, and JCS helped with data analyses. NP and JBJ wrote the final manuscript. All authors approved the final manuscript.
Neha Potnis, Ksenia Krasileva, Ralf Koebnik, João C Setubal, Brian J Staskawicz and Jeffrey B Jones contributed equally to this work.
Electronic supplementary material
12864_2010_9998_MOESM1_ESM.DOC
Additional file 1:Table S1: General features of the sequencing data and of the de novo assemblies of draft genomes of Xv, Xp and Xg using individual sequencing methods. (DOC 48 KB)
12864_2010_9998_MOESM2_ESM.PDF
Additional file 2:Figure S2: 1a) Phylogenetic tree based on MUMi indices; 1b) Distance matrix based on MUMi indices. MUMi program was used to calculate pairwise distances between draft genomes and reference Xanthomonas genomes. (PDF 93 KB)
12864_2010_9998_MOESM3_ESM.DOC
Additional file 3:Table S3: Whole genome comparisons using MUMmer dnadiff program. % coverage of the aligned contigs and % identities of the respective contigs against reference genomes has been shown for each draft genome. (DOC 36 KB)
12864_2010_9998_MOESM4_ESM.TIFF
Additional file 4:Figure S4: AvrBs2-based HR assay confirms translocation of novel effectors. Hypersensitive response reaction indicating presence of translocation signal was recorded 24 hrs after inoculation on pepper cv. ECW20R with candidate effectors xopZ2 (a), avrBs1 (b), xopG (d), xopAM (e), xopAO (f) conjugated in race 6 strain along with control race 6 strain (c). All the strains showed water-soaking on pepper cv. ECW after 48 hrs after inoculation (TIFF 946 KB)
12864_2010_9998_MOESM7_ESM.PDF
Additional file 7:Table S7: Domain architecture and distribution of proteins with HD-GYP, GGDEF and/or EAL domains encoded by genomes of different Xanthomonas strains. (PDF 348 KB)
12864_2010_9998_MOESM10_ESM.XLSX
Additional file 10:Table S10: Genes unique to Xg. Genes of special interest are highlighted in red and yellow. (XLSX 36 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Potnis, N., Krasileva, K., Chow, V. et al. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12, 146 (2011). https://doi.org/10.1186/1471-2164-12-146
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-12-146