Abstract
Background
Marbling (intramuscular fat) is a valuable trait that impacts on meat quality and an important factor determining price of beef in the Korean beef market. Animals that are destined for this high marbling market are fed a high concentrate ration for approximately 30 months in the Korean finishing farms. However, this feeding strategy leads to inefficiencies and excessive fat production. This study aimed to identify candidate genes and pathways associated with intramuscular fat deposition on highly divergent marbling phenotypes in adult Hanwoo cattle.
Results
Bovine genome array analysis was conducted to detect differentially expressed genes (DEGs) in m. longissimus with divergent marbling phenotype (marbling score 2 to 7). Three data-processing methods (MAS5.0, GCRMA and RMA) were used to test for differential expression (DE). Statistical analysis identified 21 significant transcripts from at least two data-processing methods (P < 0.01). All 21 differentially expressed genes were validated by real-time PCR. Results showed a high concordance in the gene expression fold change between the microarrays and the real time PCR data. Gene Ontology (GO) and pathway analysis demonstrated that some genes (ADAMTS4, CYP51A and SQLE) over expressed in high marbled animals are involved in a protein catabolic process and a cholesterol biosynthesis process. In addition, pathway analysis also revealed that ADAMTS4 is activated by three regulators (IL-17A, TNFα and TGFβ1). QRT-PCR was used to investigate gene expression of these regulators in muscle with divergent intramuscular fat contents. The results demonstrate that ADAMTS4 and TGFβ1 are associated with increasing marbling fat. An ADAMTS4/TGFβ1 pathway seems to be associated with the phenotypic differences between high and low marbled groups.
Conclusions
Marbling differences are possibly a function of complex signaling pathway interactions between muscle and fat. These results suggest that ADAMTS4, which is involved in connective tissue degradation, could play a role in an important biological pathway for building up marbling in cattle. Moreover, ADAMTS4 and TGFβ 1could potentially be used as an early biological marker for marbling fat content in the early stages of growth.
Similar content being viewed by others
Background
Intramuscular fat deposition in cattle starts to become visible at 12 months of age and the rate of deposition increases from 15 months to 24 months [1]. The initial formation of visible intramuscular fat seems to be driven through the development of adipocytes in combination with declining muscle growth [2]. It has been shown that marbling fat content is negatively correlated with protein content in beef muscle [3]. In addition the development of adipose tissues in longissimus muscle of high-marbled cattle appears to disorganize the structure of the intramuscular connective tissue during growth [1]. This suggests that there might be an interaction between fat development and collagen structure in muscle. Kokta et al[4] reviewed the interaction between myogenic cells and adipocytes to determine the rate and extent of myogenesis and adipogenesis during animal growth. Fat and muscle development are regulated by a number of complicated biological pathways which are related to adenoreceptor signaling [5], the cytokine signaling pathway [6] and a wide range of hormonal and transcriptional factors such as leptin [7], adiponectin [8] and insulin like growth factor protein families [9]. As such, this interaction between muscle and fat also reflects a biochemical signaling pathway within the muscle. Therefore, marbling differences might be a function of a series of complex interactions between biological pathways [10, 11].
The completion of the bovine genome project provided a tool for genome-wide functional studies to understand the interactions of complex biochemical pathways involved in protein and fat synthesis. For example, Affymetrix produces an oligonucleotide Bovine Genome Array that allows genome wide global profiling of over 23,000 bovine transcripts simultaneously Microarray based gene expression analyses associated with beef meat quality have focused on detecting differentially expressed genes in different breeds [12] and different nutritional treatments [13]. However, there is no report on gene expression differences in muscle with divergent marbling phenotypes within breed [14].
Here we report the results of a study undertaken to identify the biochemical differences in m. longissimus with divergent marbling phenotypes. The objective of this study was to identify differentially expressed genes and their role in a signaling pathway in m. longissimus with a wide range of marbling phenotypes.
Results
Differentially expressed genes between high and low marbling muscle
To detect the relationship between differentially expressed genes with marbling score, samples from m. longissimus were taken from ten unrelated animals with the highest and lowest marbling score. It was recognized that given the divergent nature of the animals the relationship would be inflated above that found in a normally sampled population.
Table 1 shows the summary statistics for the ten animals used in this microarray analysis. Intramuscular fat content (IMF) values range from 4% to 32% in the chosen animals with a clear contrast between the high and low marbled groups. After hybridization with the Affymetrix bovine genome array, gene expression intensities were measured using three pre-processing methods: MAS5.0, RMA and GCRMA.
Table 2 shows the summary of annotated probe sets on the bovine genome array. The array contains 24,128 probe sets, 91% of which were annotated by the manufacturer. Out of these, 12,745 (50%) were consistently detected as "Present (P)" in all samples according to the MAS5.0 background correction algorithm. Of the 12,745 probes in the P set, 12,184 have well annotated information (Table 2).
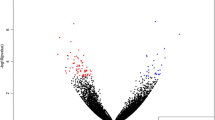
A moderated t-test using Limma (33) was used to explore genes differentially expressed between high and low marbled animals in m. longissimus., We found 136 differentially expressed genes between high and low-marbled muscle using three data processing methods: MAS5.0 (65 transcripts), RMA (37 transcripts) and GCRMA (28 transcripts) (Figure 1).
Twenty one genes showed significant differential expression (P < 0.01) in at least two normalization methods (Table 3). Out of the 21 differentially expressed genes (DEGs) listed in Table 3, eight differentially expressed genes were identified as up-regulated in muscle with high intramuscular fat content and the remaining 13 DEGs were down-regulated in samples with high intramuscular fat content. Putative genes were assigned to 14 out of the 21 DEGs (Table 3). The remaining 7 DEGs match gene sequences in bovine and other species with strong, moderate or weak sequence similarity but have no functional annotation assigned (Table 3). Based on the gene identities and associated functions, three up-regulated genes are involved in lipid metabolism (squalene epoxidase and cytochrome P450) and muscle metabolism (SH3 domain YSC-like 1). Three down-regulated genes (ATP binding protein, Proteasome activator subunit 4 and Thimetoligopeptidase 1) belong to functional classes involved in energy metabolism and an intracellular metabolic pathway. The remaining 7 DEGs are only hypothetical proteins or transcribed loci (single EST clones). Out of these 7 DEGs, three hypothetical proteins (LOC788205, LOC509649 and LOC777601) and two transcribed loci (Bt.19107.2.A1_at and Bt.19107.1.S1) could be worth pursuing further to elucidate their functional role in marbling. Technical validation of the 21 differentially expressed genes from the microarray experiment was confirmed by real-time PCR. The correlation of fold changes in gene expression between the arrays and PCR is shown in Figure 2. The results demonstrate a consistent gene expression pattern between both methods.
Gene Ontology (GO) and pathway analysis
DE genes were analyzed in the context of their GO biological process. Due to the incomplete annotation of the bovine genome, only 14 out of 21 differentially expressed probe sets were annotated. Top ranking biological GO terms are listed in Table 4, together with the genes associated to the GO term.
The pathway Studio v6.0 program (Ariadne Genomics, Inc) was used to identify molecular connections between the proteins encoded by the 14 annotated differentially expressed genes. The program searches through the ResNet database for all known interactions between genes/proteins such as physical interactions and regulation of expression (Figure 3). Out of the 14 annotated genes, 5 main pathway "hubs" (SH3KBP1, THOP1, ADAMTS4, CYP51A and SQLE) were detected in the pathway analysis. The CYP51A and SQLE proteins that are up-regulated in highly marbled muscle appear to be involved in steroid biosynthesis and cholesterol metabolism. In particular, CYP51A is activated by two proteins, SP1 and SREBP1 which are common transcription factors in lipid metabolism. SH3KBP1 is involved in cell processes such as intracellular signaling cascade, oxidative stress and cell proliferation. In addition, the pathway analysis demonstrated that ADAMTS4 is activated by immune responses related to single molecules (IL-17A, TNF, NF-kB and IL-1 family) and transforming growth factor beta 1 (TGFβ1). The results suggest a biological pathway connecting CYP51A, SQLE and ADAMTS4 that has not been previously identified in bovine gene expression studies on marbling fat.
Pathway studio analysis of 21 differentially expressed genes. Out of 14 annotated genes, 9 genes were found to be involved in direct interactions. Five main pathway "hubs" (SH3KBP1, ADAMTS4, CYP51A THOP1 and SQLE) were detected in this pathway analysis. Each arrow indicates interactions between genes and a cell process pathway.
Gene expression of selected genes in the ADAMST4 pathway
GO and pathway analysis showed that ADAMTS4 is involved in protein catabolic process (GO0006516) and it is mainly activated by the immune related single molecules IL-17A, TNFα and TGFβ1. The ADAMTS4 gene has a role in proteolysis degradation of the extracellular matrix (connective tissue) in muscle. Marbling fat accumulates in the connective tissue matrix in close proximity to blood vessels. These findings suggest that ADAMTS4 might be involved in a pathway associated to phenotypic differences of marbling fat in cattle.
To determine if any of the three regulators IL-17A, TNFα and TGFβ1 were associated with the ADAMTS4 pathway, we investigated gene expression of ADAMTS4 and the three regulators (IL-17A, TNFα and TGFβ1) in muscle with divergent IMF and protein content using RT-PCR. As shown in Figure 4, expression of ADAMTS4 increases significantly as intramuscular fat content increases (P = 0.01) and muscle protein content decreases (P = 0.01). Of the regulators only TGFβ1 significantly increased expression with increasing intramuscular fat content (P = 0.03) and it tended to decrease with decreasing muscle protein content (P = 0.08). In summary, TGFβ1 and ADAMST4 are highly associated with increases in marbling fat.
Gene expression analysis of selected genes in ADAMTS4 pathway: (A) gene expression for ADAMTS4 gene in muscle with divergent IMF and protein content. (B) gene expression for three stimulators (IL17, TNFα and TGFβ1) of ADAMTS4 protein in muscle with divergent IMF and protein content. (C) Co-expression of ADAMTS4 and TGFβ1.
Discussion
Microarray analysis
A major objective of this study was to identify new biological indicators for marbling in cattle through global transcription profiling (24K Affymetrix chip). Identification of novel differentially expressed genes might allow a better understanding of the complex biochemical mechanisms of marbling in cattle. To date, microarray based gene expression analyses for beef meat quality (marbling) have focused on detecting differentially expressed genes in different breeds of cattle such as Japanese Black (Wagyu) and Holstein Friesian cattle for a wide range of marbling phenotypes [12, 15]. As expected, many lipid metabolism related genes (for example, FABP4 and SCD) were highly expressed in the Japanese Black. As Pethick [2] commented, visible intramuscular fat content is a late maturing trait in cattle. Therefore, this study attempted to investigate genes that are differentially expressed in divergent marbling phenotypes at late stages of cattle growth.
So far, a well-known biochemical finding in relation to marbling expression in muscle was presented by Jurie et al. [16], who repored that fatty acid binding protein 4 activity was strongly correlated with intramuscular fat content. In their study they compared the different genetic performance of intramuscular fat deposition between two muscle types across three breeds. In this study however, fat metabolism related genes such as FABP4, SCD and LPL in muscle with divergent marbling phenotypes in late stage cattle were not detected. A similar study done by Liu et al.[17] in pigs indicated that FABP4 was differentially expressed among muscle samples with a divergent intramuscular fat content at 70 kg but wasn't different at a later stage (110 kg body weight). These findings suggest that many lipogenic related genes, including FABP4, will be more active during early growth during which IMF deposition is more intense, rather than at later growth stages. Alternatively, FABP4 is not detected as differentially expressed because all intramuscular fat was removed from the muscle samples used in this study (see details in the methods section).
In our study, all 21 differentially expressed genes were validated by real-time PCR. As shown in Figure 2, both methods showed consistent gene expression fold changes between the high and low marbled groups.
Functional annotation of DEGs
Gene Ontology analysis and biological pathway searches were used to explore the functional annotation of the differentially expressed genes. One of the DEGs, CDC like kinase 1(CLK1) showed a negative correlation with increase in IMF content. This gene is part of the cell cycle signaling pathway. Using a rat model, Xiao et al. 2004 found that the CLK gene was also down-regulated with muscle fat during lactation. This might be due to decreasing expression of genes involved in protein synthesis through the cell cycle signaling pathway during lactation. Another gene encoding a SH3 domainYSC-like 1 was also up-regulated in animals with high IMF content. The Src homology 3 (SH3) domain is a small protein domain of about 60 amino-acid residues which was first identified as a conserved sequence in the non-catalytic part of several cytoplasmic protein tyrosine kinases. The function of the SH3 domain is not well understood. It seems to mediate the assembly of specific protein complexes via binding to proline-rich peptides. SH3 domain containing genes, sorbin and SH3 domain containing 1 (SORBS1), which are involved in insulin-stimulated glucose uptake [18] have also been reported as up-regulated in muscle of highly marbled Japanese Black Cattle (Wagyu) [19].
Gene Ontology analysis demonstrated that several genes that were more highly expressed in high marbled muscle are involved in protein catabolic and cholesterol biosynthesis processes. This was also reflected in the pathway analysis, which generated five major pathway "hubs" (SH3KBP1, THOP1, ADAMTS4, CYP51A and SQLE) for the genes more highly expressed in high marbled muscle. CYP51A and SQLE protein appear to be involved in steroid biosynthesis and cholesterol metabolism. In particular, CYP51A is activated by two transcriptional factors, SP1 and SREBP1 in lipid metabolism. More recently, Chen et al. [20] investigated gene expression of the sterol regulatory element binding transcription factor 1 (SREBP1) in muscle from differing sexes (female and male) and differing genotype within SREBP1 gene in pig. This study showed that the SREBP1 gene was highly expressed in muscle from female compared with males. In addition there were differences in expression within the genotypes showing a strong positive correlation with intramuscular fat content. The CYP51A pathway which is driven by the SREBP1 transcription factor might be one of the biological pathways associated to intramuscular fat in cattle. In this study SREBP1 was not differentially expressed in the arrays.
Pathway for the ADAMTS4 gene
Marbling fat accumulates in a connective tissue matrix in close proximity to a blood capillary network between the bundle of muscle fibres in bovine skeletal muscle [21]. Under electron microscopy, reorganization and degradation of intramuscular connective tissue is observed in highly marbled muscle [1]. The relationship between marbling fat and protein content showed a negative correlation in longissimus of cattle; for example, longissimus with high level of fat decreased composition of moisture and protein in the carcasses [3]. Microarray analysis identified that ADAMTS4 is highly expressed in highly marbling muscle. ADAMTS4 has a function of metalopetidase that degrades the extracellular matrix of connective tissue [22]. Recently, one gene of the ADAM gene family, ADAM12 was reported as overexpressed in transgenic mice exhibiting increased intramuscular adipogenesis [23]. These findings suggest that ADAMTS4 might be one of the key genes controlling the relationship between marbling fat deposition and connective tissue degradation through a complex biological pathway in skeletal muscle.
Pathway analysis (Figure 5) suggested that ADAMTS4 is activated by three regulators; IL-17A, TNFα and transforming growth factor β 1 (TGFβ1) that have not been previously considered to be associated with marbling fat. While only ADAMTS4 was detected as differentially expressed in the arrays (TNFα was excluded in the quality control step), the RT-PCR gene expression analysis found that ADAMTS4 and TGFβ1 are highly co-expressed in highly marbled muscle while these two genes have a lower expression with higher protein content (Figure 4). The TGFβ1 is known to be a member of the GDF8 family that is a major gene known to affect carcass fatness and double muscling in cattle [24]. The GDF8 gene not only affects the size of muscle but also the proportion of connective tissue within the muscle and intramuscular fat % [19]. Also, the GDF8 gene product is a growth regulator for muscle development. In cattle the mutation of this gene product causes a decrease in fat deposition and an increase in muscle mass of carcasses [25, 26]. However, this study only shows that the ADAMTS4/TGFβ1 pathway might be involved in phenotypic differences between high and low marbled cattle. Therefore, an ADAMTS4/TGFβ1 pathway could be an important biological pathway related to increase of marbling fat in bovine skeletal muscle.
ADAMTS4 pathway. The blue solid arrow with square indicates that the regulator changes the gene expression of the target gene. The dot arrow with square indicates that the regulator changes the protein activity of the target gene. The black solid arrow indicates that the regulator changes the localization of the target gene. The blue closed line indicates that the regulator inhibits gene expression of the target gene.
Conclusion
Genome wide microarray analysis was undertaken to explore novel biological pathways associated with marbling in Hanwoo (Korean cattle). In this study, microarray analysis identified 21 differentially expressed genes (DEGs) in muscle with divergent marbling phenotypes. Pathway analysis for the 21 DEGs showed 5 unique pathway hubs associated with steroid biosynthesis, cholesterol metabolism and common transcription factors in lipid metabolism. These biological pathways might represent a phenomenon occurring in muscle with highly divergent marbling phenotypes. Out of these 5 main pathways, ADAMTS4 is involved in protein catabolic process (GO0006516), which is biologically related to the connective tissue degradation that is observed in highly marbled muscle. Pathway analysis revealed that ADAMTS4 gene is activated by three regulators; IL-17A, TNFα, and transforming growth factor β 1 (TGFβ1) that have not previously been considered to be associated with marbling fat. Gene expression analysis found that ADAMTS4 and TGFβ1 are co-expressed in highly marbled muscle while these two genes have a lower expression with higher protein content. We conclude ADAMTS4 might be one of the key genes controlling the relationship between marbling fat deposition and connective tissue degradation through a complex biological pathway in skeletal muscle. Further studies will be necessary to unveil the biological function of this pathway (ADAMTS4/TGFβ1).
Methods
Animals and sampling
The two divergent groups were formed by selecting the 5 highest and the 5 lowest marbling score carcasses from a population of 90 steers of pure bred Hanwoo cattle aged between 28 and 30 months old raised under similar conditions. Intramuscular fat (IMF %) of the muscle samples were measured using chemical fat extraction procedures and crude protein was measured on extracted samples (1 to 1.5 g) using the Macro-Kjeldahl method [27]. Table 1 presents summary statistics for all muscle samples used in this study.
Total RNA isolation and Microarray hybridization
From each animal, after removal of intramuscular fat from the samples, total RNA samples were prepared from 1 g of frozen m. longissimus tissue using Trizol reagent (Invitrogen Inc., USA) according to the manufacturer's instructions. Messenger RNA was isolated from 500 μg of total RNA using Qiagen Oligotex resin (Qiagen Inc, USA). The quantity and quality of RNA samples were measured using absorbance at 260 nm and 280 nm in a capillary type spectrophotometer (Agilent Inc, USA) and confirmed in 1.2% formaldehyde-contained gel electrophoresis.
Target preparation and high-density array hybridization
Double stranded cDNA was synthesized from 3 μg mRNA using a Genechip Expression 3'-Amplification One Cycle Synthesis kit (Affymetrix Inc. USA). The cDNA was purified using a Genechip Sample Cleanup Module (Affymetrix Inc). Biotin-labeled cRNA was synthesized in vitro using the Gene chip Expression 3'-Amplification reagents in the IVT labeling kit (Affymetrix Inc.). Biotin-labeled antisense cRNA was purified using the Genechip Sample Cleanup Module (Affymetrix Inc.) and the cDNA was fragmented in the 5 × Fragmentation buffer provided with the Genechip Sample Cleanup Module (Affymetrix Inc). Hybridization cocktail (200 μl) containing 15 μg fragmented cRNA was injected into the Genechip Bovine Genome Array (Affymetrix Inc). The array was placed in a 45 degrees hybridization oven at 60 rpm for 16 hours. After hybridization, the arrays were washed and stained in a fluidic station with the appropriate signal amplification protocol using biotinylated anti-streptavidin antibody (Vector Laboratories Inc., USA) and phycoerythrin-conjugated streptavidin (Invitrogen Inc., Carlsbad, CA, USA). The array was scanned with a GeneChipScanner 3000 (Affymetrix Inc.).
Microarray Data processing
All quality control measures, preprocessing and analyses were performed using the statistical computing language R [28]. The quality of the arrays was assessed through standard quality control measures for Affymetrix arrays: pseudoimages of the arrays, MA scatter plots of the arrays versus a pseudomedian reference chip and other summary statistics including histograms and box plots of raw log intensities, box plots of relative log expressions, box plots of normalized unscaled standard errors and RNA degradation plots [29]. Transcription intensities in log2 scale were estimated from the probe-level data by using three summarization methods: MAS5.0 [30], RMA [30] and GCRMA [31]. In MAS5.0, each probe was adjusted using a weighted average. All arrays were scaled to the same mean value for normalization (200) and were summarized by a log2 scale average using 1-step Tukey biweight. For RMA, the background was corrected by convolution. The data were quantile normalized and summarized by median polish. GCRMA background correction used an affinity measure model based on probe sequences and mismatch intensities. Presence calls [32] for the probes were also calculated (τ = 0.015, α1 = 0.04 and α2 = 0.06). The data were filtered to remove control probes (n = 133) and probes detected as marginal or absent in all arrays using MAS5 presence calls.
Microarray data analysis
Prior to testing for differential expression, the data were filtered to remove Affymetrix control probes (n = 133) and all noninformative probes detected as marginal or absent in all arrays (n = 11,383), thus remaining 12,745 probes to be tested. Differential transcription was tested for each summarization method using LIMMA [33, 34]. Only differentially expressed (DE) probes detected in two out of the three summarization methods (P < 0.01) and flagged as present in at least 50% of the samples were considered to be significant. This approach ensures maximum specificity to detect differential expression and minimizes the effect of different summarization methods which are the main source of variability in the analysis of Affymetrix arrays. No false discovery rate correction method is warranted due to the stringency of the filtering criteria [35].
QRT-PCR for the 21 DEGs and selected genes from the ADAMTS4 pathway
For technical validation of the microarray results, the 21 differentially expressed genes were validated by quantitative realtime RT-PCR (qRT-PCR). cDNA synthesis was performed using 2 μg of total RNA. The complimentary strand was primed with a random primer (Promega.co) and cDNA synthesis was performed using a Superscript III kit (Invitrogen) according to the manufacturer's instructions. Each quantitative PCR was carried out in a final volume of 20 containing 1 μl cDNA (500 ng/μl), 2 × SYBR Green I Master Mix (10 μl) (Qiagen., GmbH, Germany), and 10 pM forward and reverse primers. The real-time PCR reactions started at 95°C for 15 min for pre-denaturation and the condition was set at 95°C for 10 s, 56°C for 20 s and 72°C for 30 s. The PCR performed 40 cycles. The PCR was conducted in ABI 7500 realtime PCR system (Applied Biosystems, USA). Primer sets used in the real-time PCR are listed in Table 5.
The relative gene expression value was calculated by the ΔCt method [36]. The ΔCt value of the target gene was normalized against the G3PDH Ct value. The fold change was determined as 2-ΔCt.
For selected genes in the putative ADAMTS4 pathway, a regression analysis of gene expression value (2-ΔCt) on intramuscular fat and protein content was performed using a simple linear regression in R [28].
Gene ontology (GO) analysis
Annotation of DE probes was performed using the Database for Annotation. Visualization and Integrated Discovery (DAVID) http://david.abcc.ncifcrf.gov/home.jsp. In subsequent text the term "probe" is replaced by "gene". The DE genes were analyzed in the context of their gene ontology (GO) biological process (Gene Ontology Consortium. 2006).
Biological pathway analysis
Differentially expressed genes in the m. longissimus of high and low marbled Hanwoo were clustered into pathways using the program Pathway studio (Stratagene, La Jolla, CA). This program provided a visual representation of the differentially expressed genes from the microarray data. In this study, 14 annotated genes out the 21 differentially expressed genes were imported into the program. Pathway studio then generated relationships between gene products using a literature search of curated genes and displayed the interactions in the form of pathways that can also include other genes/proteins, small molecules, cellular processes, and relevant transcription factors. If no interactions were found or proper annotation was not available, the gene was not included in the pathway. To help explicit the function of the genes in each pathway, gene ontology terms were determined for all genes illustrated within the pathways generated by Pathway studio 6.0.
References
Nishimura T, Hattori A, Takahashi K: Structural changes in intramuscular connective tissue during the fattening of Japanese black cattle: effect of marbling on beef tenderization. J Anim Sci. 1999, 77: 93-104.
Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C, Pethick DW: Intramuscular fat content in meat-producing animals:development, genetic and nutritional control, and identification of putative markers. Animal. 2010, 4: 303-319. 10.1017/S1751731109991091.
Brackebusch SA, McKeith FK, Carr TR, McLaren DG: Relationship between longissimus composition and the composition of other major muscles of the beef carcass. J Anim Sci. 1991, 69: 631-640.
Kokta TA, Dodson MV, Gertler A, Hill RA: Intercellular signaling between adipose tissue and muscle tissue. Domestic Animal Endocrinology. 2004, 27: 303-331. 10.1016/j.domaniend.2004.05.004.
Fruhbeck G, Gomez-Ambrosi J, Muruzabal FJ, Burrel MA: The adipocyte: a model of integration of endocrine and metabolic signaling in energy metabolism regulation. Am J Physiol. 2001, 280: E820-847.
Shin SM, Kim K, Kim JK, Yoon SR, Choi I, Yang Y: Dexamethasone reverses TGF-beta-mediated inhibition of primary rat preadipocyte differentiation. FEBS Lett. 2003, 543: 25-30. 10.1016/S0014-5793(03)00371-5.
Lissett C, Clayton PE, Shalet S: The acute leptin response to GH. J Clin Endocrinol Metab. 2001, 86: 4412-4415. 10.1210/jc.86.9.4412.
Wu X, Motoshima H, Mahadev K, Stalker TJ, Scalia R, Goldstein BJ: Involvement of AMP-activated protein kinase in glucose uptake stimulated by the globular domain of adiponectin in primary rat adipocytes. Diabetes. 2003, 52: 1355-1363. 10.2337/diabetes.52.6.1355.
McCusker RH: Controlling insulin-like growth factor activity and the modulation of insulin-like growth factor binding protein and receptor binding. J Dairy Sci. 1998, 81: 1790-1800. 10.3168/jds.S0022-0302(98)75748-0.
Sorisky A: From preadipocyte to adipocyte: differentiation-directed signals of insulin from the cell surface to the nucleus. Crit Rev Clin Lab Sci. 1999, 36: 1-34. 10.1080/10408369991239169.
Hausman DB, DiGirolamo M, Bartness TJ, Jausman GJ, Martin RJ: The biology of white adipocyte proliferation. Obes Rev. 2001, 2: 239-254. 10.1046/j.1467-789X.2001.00042.x.
Wang YH, Reverter A, Mannen H, Taniguchi M, Harper GS, Oyama K, Byrne KA, Oka A, Tsuji S, Lehnert SA: Transcriptional profiling of muscle tissue in growing Japanese Black cattle to identify genes involved with the development of intramuscular fat. Aust J Exp Agric. 2005, 45: 809-820. 10.1071/EA05058.
Byrne KA, Wang YH, Lehnert SA, Harper GS, McWilliam SM, Bruce HL, Reverter A: Gene expression profiling of muscle tissue in Brahman steers during nutritional restriction. J Anim Sci. 2005, 83: 1-12.
Bernard C, Cassar-Malek I, Renand G, Hocquette JF: Changes in muscle gene expression related to metabolism according to growth potential in young bulls. Meat Science. 2009, 82: 205-212. 10.1016/j.meatsci.2009.01.012.
Wang YH, Byrne KA, Reverter A, Harper GS, Taniguchi M, McWilliam SM, Mannen H, Oyama K, Lehnert SA: Transcriptional profiling of skeletal muscle tissue from two breeds of cattle. Mammalian Genome. 2005, 16: 201-210. 10.1007/s00335-004-2419-8.
Jurie C, Cassar-Malek I, Leroux C, Bauchart D, Boulesteix P, Pethick DW, Hocquette JF: Adipocyte fatty acid-binding protein and mitochondrial enzyme activities in muscles as relevant indicators of marbling in cattle. Journal of Animal Science. 2007, 85: 2660-2669. 10.2527/jas.2006-837.
Liu J, Damon M, Guitton N, Guisle I, Ecolan P, Vincent A, Cherel P, Gondret F: Differentially Expressed genes in Pig Longissimus Muscles with contrasting levels of Fat, as indentitied by combined transcriptom ic, reverse transcription PCR and proteomic analyses. J Agric Food Chem. 2009, 57: 3808-3817. 10.1021/jf8033144.
Baumann CA, Ribon V, Kanzaki M, Thurmond DC, Mora S, Shigematsu S, Bickel PE, Saltiel AR: CAP defines a second signaling pathway required for insulin-stimulated glucose transport. Nature. 2000, 407: 202-207. 10.1038/35025089.
Sasaki Y, Nagai K, Nagata Y, Doronbekov K, Nishmura S, Yoshioka S, Fujita T, Shiga K, Miyake T, Taniguchi Y, Yamada T: Exploration of genes showing intramuscular fat deposition associated expression changes in musculus longissimus muscle. Animal Genetics. 2005, 37: 40-46. 10.1111/j.1365-2052.2005.01380.x.
Chen J, Yang XJ, Xia D, Chen J, Wegner J, Jiang Z, Zhao RQ: SREBF1 expression and genetic polymorphism significantly affect intramuscular fat deposition in the longissimus muscle of Erhualian and Sutai pigs. Journal of Animal Science. 2008, 86: 57-63. 10.2527/jas.2007-0066.
Harper GS, Pethick DW: How might marbling begin?. Australian Journal of Experimental Agriculture. 2004, 44: 653-662. 10.1071/EA02114.
Taniguchi Y, Takano A, Doronbekov K, Sugimoto Y, Yamada T, Sasaki Y: Genomic organization and promoter analysis of the bovine ADAM12 gene. Animal Biotechnology. 2008, 19: 178-189. 10.1080/10495390802212445.
Kawaguchi N, Sundberg C, Kveiborg M, Moghadaszadeh B, Asmar M, Dietrich N, Thodeti GK, Nielsen FC, Moller P, Mercurio AM, Albrechtsen R, Wewer UM: ADAM12 induces actin cytoskeleton and extracellular matrix reorganization during early adipocyte differentiation by regulating beta1 integrin function. J Cell Sci. 2003, 116: 3893-3904. 10.1242/jcs.00699.
Grobet L, Martin LJR, Roncelet D, Pirottin D, Browers B: A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nature Genetics. 1997, 17: 71-74. 10.1038/ng0997-71.
Hanset R, Michaux C, Dessy-Doize C, Burtonboy G: Studies on the 7th rib cut in double muscled and conventional cattle. Anatomical, histological and biochemical aspects. In Muscle hypetrophy of genetic origin and its use to improve beef production. Edited by: King JWB, Menissier F. 1982, P341-
Shahin KA, Berg RT: Fat growth and partitioning among the depots in double muscled and normal cattle. Canadian journal of animal science. 1985, 65: 295-306. 10.4141/cjas85-036.
AOAC: Official Methods of Analysis. 1996, Washington DC
R Development Core Team: R: a language and environment for statistical computing. 2008, R Foundation for statistical computing, Vienna, Austria
Bolstad BF, Collin B, Brettschneider J, Simpson K, Cope L, Irizarry R, Speed T: Quality assessment of Affymetrix GeneChip Data. 2005, P33-47.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Speed TP: Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003, 31: e15-10.1093/nar/gng015.
Wu Z: A model based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2003, 99: 909-917. 10.1198/016214504000000683.
Liu W, Mei R, Di X, Ryaid J, Sder T, Hubbell E, Dee S, Webster T, Harrington C, Ho M, Baid J, Smeekens S: Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002, 18: 1593-1599. 10.1093/bioinformatics/18.12.1593.
Smyth GK: Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Edited by: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. 2005, Springer, New York, 397-420. full_text.
Smyth GK: Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004, 3: 10.2202/1544-6115.1027.
Gondro C: Summarization methods and quality problems in Affymetrix microarrays. Proc Assoc Advmt Anim Breed Genet. 2009, 18: 304-307.
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using realtime quantitative PCR and the 2-∆∆Ct method. Methods. 2001, 25: 402-408. 10.1006/meth.2001.1262.
Acknowledgements
This work was supported by awards from the International Collaborative research fund (Grant No: 200712A01032083) between the Rural Development Administration (RDA) in Korea and Cooperative Research Centre for Beef Genetic Technologies in Australia. Seung Hwan Lee held International Postgraduate Research Scholarship (IPRS) from University of New England (UNE). The authors also thank the anonymous reviewers for their helpful suggestions and comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
S.H.L and C.G were involved in designing and planning of the study. They carried out the experiments and analyses, interpreted data, annotation and statistical analyses and wrote the first draft of the paper. N.K.K and D.L were involved in the realtime PCR analysis, pathway analysis and contributed to the writing of the paper. E.W.P and S.J.O was involved in the experimental design and contributed to the writing of the paper. S.J.O, J.V, J.P.G and J.M.T conceived the project, designed and carried out analysis, interpreted data, and were involved in the writing of the paper. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lee, SH., Gondro, C., van der Werf, J. et al. Use of a bovine genome array to identify new biological pathways for beef marbling in Hanwoo (Korean Cattle). BMC Genomics 11, 623 (2010). https://doi.org/10.1186/1471-2164-11-623
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-11-623