Abstract
Background
Genetically anchored physical maps of large eukaryotic genomes have proven useful both for their intrinsic merit and as an adjunct to genome sequencing. Cultivated tetraploid cottons, Gossypium hirsutum and G. barbadense, share a common ancestor formed by a merger of the A and D genomes about 1-2 million years ago. Toward the long-term goal of characterizing the spectrum of diversity among cotton genomes, the worldwide cotton community has prioritized the D genome progenitor Gossypium raimondii for complete sequencing.
Results
A whole genome physical map of G. raimondii, the putative D genome ancestral species of tetraploid cottons was assembled, integrating genetically-anchored overgo hybridization probes, agarose based fingerprints and 'high information content fingerprinting' (HICF). A total of 13,662 BAC-end sequences and 2,828 DNA probes were used in genetically anchoring 1585 contigs to a cotton consensus genetic map, and 370 and 438 contigs, respectively to Arabidopsis thaliana (AT) and Vitis vinifera (VV) whole genome sequences.
Conclusion
Several lines of evidence suggest that the G. raimondii genome is comprised of two qualitatively different components. Much of the gene rich component is aligned to the Arabidopsis and Vitis vinifera genomes and shows promise for utilizing translational genomic approaches in understanding this important genome and its resident genes. The integrated genetic-physical map is of value both in assembling and validating a planned reference sequence.
Similar content being viewed by others
Background
The Gossypium (cotton) genus, composed of 50 species among which four provide the major raw material for one of the world's largest industries (textiles), has a large impact on our economy and everyday life. Diploid cottons are classified into 8 genome types, denoted A-G and K, based on chromosome pairing relationships [1]. All diploid cotton species are believed to have shared a common ancestor about 5-10 million years ago [1]. The cotton genome types diverged into genome groups that vary in haploid genome size from 2500 Mb in the K genome, to less than 900 Mb in the D genome [2, 3], while retaining common chromosome number (n = 13) and largely-collinear gene order [4–7]. The tetraploid cotton genome is thought to have formed by an allopolyploidy event about 1-2 million years ago, involving species similar to the modern New World D genome species G. raimondii (GR) [8] or G. gossypioides (GG) [9] and the Old World A genome species G. herbaceum (GH).
There exist at least a dozen published genetic maps for various Gossypium crosses, most involving members of the superior-fiber-quality G. barbadense species crossed with high-yielding G. hirsutum. These maps collectively include > 5,000 public DNA markers (~3,300 RFLP, 700 AFLP, >2,000 SSR, and 100 SNP). Many thousands of additional SSRs have been described [10], but only a subset of these have been mapped [4, 11–13]. The most detailed sequence tagged site (STS)-based map, and a source of probes for many of the other maps, are reference genetic maps for diploid (D) and tetraploid (AtDt, where Dt refers to the D-subgenome found in tetraploid cottons (to distinguish it from the genome of D-diploid cottons). Likewise, At refers to the A-subgenome of tetraploid cottons.) Gossypium genomes that include respectively, 2584 loci at 1.72 cM (~600 kb) intervals based on 2007 probes (AtDt); and 1014 loci at 1.42 cM (~600 kb) intervals detected by 809 probes (D) [4, 14]. A high degree of collinearity among the respective genome types permitted inference of the gene order of a hypothetical common ancestor of the At, Dt, and D genomes for 3016 loci identified by 2337 probes, spanning 2324.7 cM [14]. Additional maps that are particularly marker-rich and/or have been widely used as reference maps for QTL studies have been developed from three additional interspecific crosses [11–13]. Other important resources include aneuploid substitution stocks that were derived from tetraploid genotypes TM-1 (G. hirsutum) × 3-79 (G. barbadense) [15] and TM-1 × G. tomentosum[16]. Together, monosomics and telosomics have been used to assign 20 of the 26 cotton linkage groups to chromosomes, and the remaining six linkage groups were assigned to chromosomes by translocation and fluorescence in situ hybridization mapping [17].
Cotton genetic maps have been employed in identification of diagnostic DNA markers for a wide range of traits related to fiber yield and quality [18–46]; drought tolerance [46–48]; and resistance to diseases [49–54], and pests [55–59]. Interest in hybrid cottons in some countries has drawn attention to a nuclear restorer of cytoplasmic male sterility [60–64]. Morphological features such as the pubescence that is characteristic of G. hirsutum[65–68], leaf morphology [69–72] and color [73], and unique features such as nectarilessness [34, 69, 74] have also received attention. The value of cotton seed has led to interest in mapping variation in seed physical characteristics and nutritional value [75]. Meta-analysis of multiple QTL mapping experiments by alignment to a common reference map has begun to reveal the genomic organization of trait variation [76]. Although members of the D genome clade do not make spinnable fiber, genetic mapping has shown that the majority of fiber QTLs mapped in tetraploid cotton fall on D genome (G. raimondii-derived) chromosomes, suggesting that the D genome has been crucial to the evolution of the higher fiber quality and yield of cultivated tetraploid cottons [76].
Toward the long-term goal of characterizing the spectrum of diversity among the 8 Gossypium genome types and three polyploid clades, the worldwide cotton community has prioritized the D-genome species Gossypium raimondii for complete sequencing [77, 78]. Gossypium raimondii is a diploid with a ~880 Mb genome [3], the smallest genome in the Gossypium genus at ~60% of the size of the diploid A genome and 40% of the tetraploids. It is largely inbreeding, and a largely-homozygous genotype has been used in both a reference genetic map [4] and for a BAC library (herein). DNA renaturation kinetics shows that 30-32% of the G. raimondii genome contains repetitive DNA, with a kinetic complexity of 1.6 × 106 bp and an average iteration frequency of ~120 copies per haploid genome [79]. This has been subdivided into a highly-repetitive component of about 5% of the genome, composed of elements in 10,000 or more copies; and a middle-repetitive component accounting for 27% of the genome [80]. A random sampling of 0.04% of the tetraploid cotton genome, enough to sample repetitive element families that occur in 2500 or more copies, revealed only 4 D-genome-derived elements ranging in estimated copy number up to about 15,000, versus dozens of A-genome-derived repeats at much higher copy numbers [81]. Pilot sequencing studies (X. Wang, D. Rokhsar, A.H. Paterson, unpubl.) show that most D-genome repetitive DNA families are sufficiently heterogeneous to be compatible with a whole-genome shotgun approach.
Genetically anchored physical maps of large eukaryotic genomes have proven useful both for their intrinsic merit and as an adjunct to genome sequencing. In species where no whole-genome sequence is yet available, a physical map is a useful tool in a wide range of activities including comparative genomics and gene cloning. Physical mapping also provides a method of genome assembly independent of a sequence, and is useful in contributing to and/or validating whole-genome shotgun sequences [82]. For BAC-based sequencing of a genome, a physical map is a prerequisite. Recent study of chromosomes 12 and 26 of upland cotton (Gossypium hirsutum) [83] suggests that physical mapping of polyploid cotton may be complicated by homoeologous genome fragments.
As an important step toward its genome-wide characterization, we describe here a genetically anchored, BAC-based physical map for G. raimondii. By incorporating thousands of DNA markers, the physical map is tightly integrated with the rich history of cotton molecular genetics research described above, and expedites a host of studies of Gossypium biology and evolution. Moreover, comparison of the physical map to the sequences of Arabidopsis thaliana and Vitis vinifera shows promise for utilizing translational genomic approaches in better understanding the structure, function, and evolution of this important genome and its resident genes.
Results
BAC library
The Gossypium raimondii BAC library used in physical mapping consists of 92,160 clones. Pulsed-field gel electrophoresis-based examination of 448 Not I digested clones indicates a mean insert size of 100 kb. Of note, there was little variation in insert size among clones (standard error of mean = 0.76). Three of the 448 interpretable Not I-digested clones (i.e., 0.67%) appear to be false positives. Likewise, three of the 4032 BAC end sequences generated from the library exhibit homology to chloroplast DNA (0.07%) indicating that the methods employed in constructing the library [84] were successful in keeping chloroplast contamination low. Collectively, the library affords 10× coverage of the G. raimondii genome.
Agarose-based fingerprints and HICF
Two different types of fingerprints were employed in this study: a preliminary assembly used agarose-based fingerprinting and an improved assembly resulted from re-fingerprinting a subset of BACs using HICF. The entire 92,160-clone GR BAC library was fingerprinted using slight modification of established agarose-based fingerprinting methods [85]. Preliminary assembly formed 9,290 contigs and 26,716 singletons at a tolerance value of 8 and cutoff value of 1e-10. The average agarose-based fingerprint band number of individual BACs was 17.4. Band number distribution across the library is shown in Figure 1A. A total of 3266 BACs failed to produce usable fingerprints.
Comparison of band number distribution between agarose-based fingerprints and HICF. A: agarose-based fingerprints; B: HICF; C: an example of two agarose FPC contig joined in HICF. Red bands are matching bands to the highlighted (in blue) BAC. Counts of matching bands to the BAC are listed below each lane. The four BACs on the right were not assembled into the same contig.
Two terminal BACs from each end of the largest 4608 agarose contigs (four BACs per contig, totaling 18,432 BACs) from the preliminary assembly were fingerprinted using HICF. The average HICF band number per BAC was initially 203.6. HICF batches with extremely high or low band numbers (approximately top or bottom 5%) were re-fingerprinted. The average band number dropped to 178. These 18,432 BACs formed 3508 contigs and 2570 singletons. The final band number distribution is shown in Figure 1B.
Overgo hybridizations
Thousands of probes were applied to the GR library using a multiplex hybridization scheme (see Methods). A total of 2828 probes from Arabidopsis genes, cotton ESTs, and genetic markers showed hybridization signal attributable to one or more BACs by this approach. On average, each probe hit 17.3 BACs. A total of 46 probes hit more than 100 BACs and are considered highly repetitive. To minimize false associations, probes with >50 hits were not used in the contig assembly process, and probes with >30 hits were not used in the contig anchoring process (detailed later). Thus, 2658 probes (with <50 hits) were integrated into the assembly using the CpM table in FPC: stringency (cutoff value) was relaxed by 2, 3, or 4 denary (ten-fold) intervals when 1, 2 and 3+ common markers were found between two BACs.
Integrated assembly
Since agarose-based fingerprinting and HICF use different sets of restriction enzymes, a different band-calling scheme, and have different error rates and band size tolerances, data from these two different methods cannot be merged directly. Further, while we targeted HICF to contig-terminal BACs, it would be imprudent to declare a join in the agarose assembly whenever HICF suggests a merge of contig-terminal BACs, overlooking potential false joins in HICF. To circumvent this, if two agarose contig-terminal BACs were suggested to be joined by HICF, we lowered the cutoff value for joining agarose contig-terminal BACs by two denary intervals, e. g. when the overall cutoff was set to 1e-12, we would accept an overlap at the cutoff at 1e-10 if the two BACs were found in the same HICF contig. The agarose assembly was thus reassembled, only forming a merged contig if it was supported by both data types (see Methods), and integrating 2658 hybridization markers based on 2828 overgos.
Collectively, the agarose fingerprints, targeted HICF fingerprints, and overgo hybridization data joined a total of 67,343 BACs into 4208 contigs, leaving 21,551 singletons. Based on the average insert size estimate of 100 kb, and an estimated genome size of 880 Mbp [3], the 67,343 BACs in contigs provide ~7.7× coverage of the GR genome. The majority of contigs (61.5%) contain between 3 and 25 BACs. The distribution of BAC numbers per contig is shown in additional file 1.
Singletons differed in several ways from BACs in contigs. The average agarose-based fingerprint band number was 13.4 for singletons, versus 17.9 for BACs in contigs. A total of 9476 (44% of) singletons contained less than 12 bands. This could reflect either shorter length of singleton BACs, or the presence of tandem repeats that produce fingerprint bands that comigrate, reducing the scoreable band number and perhaps contributing to failure of some BACs to form contigs (see more discussion of band numbers below). A total of 1904 overgo probes hit singleton BACs, among which 364 overgos were repetitive and 1540 were low copy (having <30 hits total). Compared to the probes that hit BACs in contigs (376 repetitive and 2129 low copy), singletons show some enrichment in repetitive DNA content. A total of 585 singletons were identified as possible cross-well contaminations.
Anchoring contigs to the cotton consensus map
After filtering out 381 (of 2828) repetitive overgo probes that hit more than 30 BACs in the GR library, and 357 BACs (out of 34,713 BACs with at least one marker hit) with more than 8 markers hybridized as suspected hybridization artifacts, the remaining probes and BACs produced 40,152 BAC-probe pairs. A total of 7772 of these were produced by BACs that were not in contigs (singletons); 5946 of the markers on contigs were "weak anchors" produced by a single BAC-probe pair for the contig. Weak anchors were not used in aligning the contigs onto the genetic map. The remaining 26,434 BAC-probe pairs derive from 1920 probes, and were distributed in 2154 contigs.
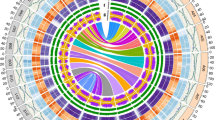
A 'consensus' cotton genetic map built from the At, Dt and D genome genetic maps contains 13 homologous groups made up of 3016 loci based on 2337 unique sequence tags [14]. Among these, 2109 have probes designed (961 RFLP probes and 1744 overgos, 596 have both, most of the remainder could not be sequenced). After filtering out probes with >30 hits in the library, 1468 loci on the consensus map have anchored 1586 contigs. (Table 1, Figure 2, S1). On average, each marker anchored 2.42 contigs.
Aligned physical map contigs along the consensus map. Homologous Group 1 of the integrated genetic-physical map, drawn using data from Rong et al. [14].
Aligning contigs to Arabidopsis thaliana and Vitis vinifera whole-genome sequences
A total of 8064 BACs selected from the ends of the largest 2016 contigs from the preliminary assembly were used for paired-end sequencing. The resulting 13,662 high-quality sequences, along with the 1920 low copy probes (after filtering described above), were used in comparing the GR contigs to Arabidopsis thaliana (AT) and Vitis vinifera (VV) chromosomes.
BAC end-sequences (BES) and the source sequences of the hybridization probes were aligned to the AT and VV whole-genome sequences using BLASTn. A total of 2607 sequences (1370 BES and 1237 overgo source sequences) had between 1 and 9 BLAST hits in the AT genome, and 2968 sequences (1557 BES and 1411 overgo source sequences) have between 1 and 9 hits in the VV genome. (Sequences with >10 hits were excluded as repetitive.)
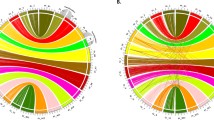
A total of 370 contigs were aligned to Arabidopsis chromosomes, 438 to Vitis chromosomes, and 242 to both (Table 2, Figure 3). All 566 that aligned contained 64 CB units (consensus band units, the number of total non-overlapping bands in a contig) per contig on average, about 50% larger than the overall average contig size (42 CB units). Based on an estimated size of 4097 bps per band (average of all band sizes from all BACs fingerprinted), these contigs cover a minimum of 13% (contigs anchored on VV) and 11% (contigs anchored on AT) of the GR genome, noting that band numbers somewhat underestimate contig sizes because both very large and very small bands are excluded from bandcalling. A second estimate of coverage of the target genomes by aligned contigs was obtained by adding up the distances between anchor marker BLAST matches and excluding overlaps. This suggests that 27.7% of the Arabidopsis genome and 22.8% of the Vitis genome is covered by aligned GR contigs. Some contigs have significant association with two or more positions on a target genome. The distributions of contigs along AT and VV chromosomes are shown in Figure 3. Contigs are more likely to be anchored to two or more locations in AT than VV (159 or 43% of contigs anchor to multiple AT locations versus 111 or 25.4% of contigs anchored to VV), consistent with the fact that the Arabidopsis lineage has experienced two more whole-genome duplication (WGD) events than grape [86].
Aligned GR contigs along Arabidopsis and Vitis chromosomes. Blue and purple bars next to the chromosomes show the GR contigs. Green lines to the left of each chromosome indicate gene-density of the target genomes. The length of the bar represents the physical distance between anchoring markers on the target genomes. Putative centromeric regions on Arabidopsis chromosomes are marked out in rectangles. Centromeric regions on Vitis chromosomes cannot be determined.
The GR contigs anchored on VV are not evenly distributed across the chromosomes, but rather are clustered in several regions/chromosome arms that tend to have higher than average gene densities. Gene density distribution across the Vitis genome was extracted by counting the number of genes in 200 kb bins along the chromosomes. Gene density is largely uniform across the Arabidopsis chromosomes except for the centromeric regions; while in the Vitis genome, we observed greater heterogeneity of gene density. The regions on which we were able to anchor GR contigs (Figure 3) had an average of 20 genes per 200 kb window, versus an average of 14.8 for the remainder of the genome. Among the 30% of VV 'windows' with highest gene density, 37.9% were covered by GR contigs; versus 22.8% of the genome as a whole.
Nature of repetitive probes
A total of 46 probes are classified as highly-repetitive (with >100 BAC hits). These came from several sources: 28 were derived from cotton EST sequences (COV), 3 were derived from genes that are low-copy in Arabidopsis (AOG), and 15 were derived from cotton RFLP probes used in genetic mapping (see additional file 2 for the complete sources of these probes). Six of the highly repetitive cotton overgo sequences were found to be located within known repetitive elements using Repbase http://www.girinst.org/repbase/. The overgo with the most hits (COV1526, which hits 1593 BACs) is in a helitron. The remaining five were from two hAT-like DNA transposons, one EnSpm element, one ERV/ERV2 element and one Gypsy element. Four of the 15 highly repetitive PCR-based probe sequences contain repetitive elements. The three Arabidopsis genes from which highly repetitive overgos were designed (At5g10360, At2g30740 and AtGRF2) showed no known repetitive elements in their sequences, which might indicate cotton lineage-specific gene multiplications. Given that Repbase does not include a comprehensive set of cotton repetitive sequences (due to lack of a complete Gossypium genome), it is likely that the remaining highly repetitive overgos that did not match repetitive sequences from Repbase may reveal cotton elements not previously known to be repetitive.
Low-copy and repetitive DNA loci were concentrated in different regions of the genome
A total of 3060 contigs contain BACs to which one or more probes hybridized. Probes were classified as low copy (<30 hits total), moderately repetitive (31-97 hits), or highly repetitive (>100 hits). Accordingly, contigs were tentatively classified as repetitive or low-copy based on the ratio of repetitive probes versus low-copy probes hybridized to each contig. A total of 761 contigs contain only repetitive probes, and 1262 contigs contains mostly (>60%) low copy probes. Because a large number of the probes are designed from cotton EST sequences or Arabidopsis genes, contigs with relatively more hybridization anchors from low copy probes and relatively fewer from repetitive probes are likely to be gene rich. The 1262 low-copy probe enriched contigs contain 1786 of the 2300 non-repetitive probes. The majority of the low-copy probe enriched contigs (901 out of 1262, or 71.4%) are anchored to the cotton consensus map (additional file 3). By comparison, only 37.7% (1586 out of 4208) of contigs overall could be anchored to the consensus map.
Repetitive contigs are slightly shorter than contigs enriched in low copy probes (average 38.32 CB units versus 44.35 CB units). This could be caused by co-migrating fragments produced by the repetitive sequences that reduce the total number of bands.
Low-copy probe enriched contigs appear to be largely euchromatic
Among the 438 contigs that showed microsynteny to VV chromosomes, 218 are enriched in low-copy probes and only 14 are repetitive probe enriched. Similarly, among the 370 contigs that showed microsynteny to AT chromosomes, 166 were enriched in low copy probes, and only 17 are repeat-enriched. This is consistent with our prior findings in other taxa that microsynteny tends to be preserved in gene-rich euchromatic regions but not in repeat-rich heterochromatic regions [87]. We tacitly assume that the 761 repeat-enriched contigs are likely to be largely from heterochromatic regions of the genome and the 1262 low-copy sequence-enriched contigs are likely to be from euchromatic regions of the genome. The 1262 low-copy contigs can be estimated to cover 26% of the genome based on the estimated genome size of 880 M and average band size of 4097 bp. Based on the 68% of the genome estimated to be low-copy by renaturation kinetics[79], these contigs may cover about 38.2% of the low-copy DNA. Contigs aligned to VV and AT genomes contains 1150 (50%) and 954 (41.5%) of all non-repetitive probes. The low copy probes that were unable to align were partly due to the limitation of BLAST in searching across distant related species and the variation in gene density in VV genome.
Consequences of ancient duplications in the Arabidopsis thaliana genome
To illustrate the alignment of GR contigs on the AT and VV genomes, ctg500 was used as an example. The contig is anchored to a single VV chromosomal location at about 14.7 Mb on chr8, and to four different locations on the AT genome, at 15 Mb on chr2, 2.7 Mb on chr3, 20 Mb on chr3 and 0.1 Mb on chr5 respectively (Figure 4A). These four AT regions were previously shown to be paralogous segments created by two rounds of whole-genome duplication [88]. The chromosomal region in Vitis has also been identified using MCScan [89], to have conserved collinearity with the four AT regions (Figure 4B). Ctg500 is anchored on cotton consensus homologous group 2, at around 67 cM. Based on cotton DNA markers, this region has shown evidence of homology to Arabidopsis α11 and α14 groups [14].
Alignment of contig 500 to the genome sequences. A. The contig is mapped to four regions in Arabidopsis, which are paralogs produced by the α and β duplications after the cotton-Arabidopsis divergence. The contigs are only anchored to a single Vitis chromosomal location. B. dot plot generated by MCscan on Plant Genome Duplication Database, showing conserved syntenic blocks between Vitis chr.8 and Arabidopsis chromosomes. The region corresponding to GR ctg 500 is marked by red circles.
The G. raimondii chloroplast
By aligning to the chloroplast DNA sequence of upland cotton (Gossypium hirsutum) using BLAST, BAC-end sequences and probes likely to be of chloroplast origin were identified. Ctg11556 is identified as a chloroplast contig. The contig contains 20 BACs, 10 of which are "buried" in FPC, meaning they have nearly identical band patterns as other BACs in the contig, indicating very high similarity among these BACs. COV1960, an overgo probe designed from the sequence of the chloroplast psaJ gene, hits 17 of the 20 BACs in the contig. Three BACs from the contig have end sequences, all of which correspond to the published G. hirsutum chloroplast sequence (additional file 4). Based on low-coverage genomic sequencing with some targeted finishing, a D-genome chloroplast sequence has been assembled and is being described (M. Rahman, A. H. Paterson, in prep.)
GO analysis of BES and shotgun sequences
The 13,662 BES were analyzed using Blast2Go to obtain a distribution of functional gene groups. A total of 9042 did not have significant hits using BLASTx against NCBI nr database, 3234 of the sequences are annotated, and 963 were mapped, but not annotated. No significant differences were observed between the GO distribution of BES and random shotgun sequences except that more genes involved in localization processes were represented in the random shotgun sequences. (see additional files 5 and 6).
Discussion
The first whole-genome physical map of a cotton species has provided new tools and information, and foreshadows the picture of cotton genome organization prior to the completion of the D-genome sequencing currently in progress. The genetically anchored contigs are potentially helpful in efforts such as gene cloning and local sequence analysis, by providing region-specific BAC resources for marker development and chromosome walking. On a genomic level, comparative analysis between cotton, Arabidopsis, and Vitis genomes illustrates the potential for translational genomics across these species, and several regions with an unusually high degree of conserved collinearity may be interesting for further research.
Several lines of evidence herein suggest that the G. raimondii genome is comprised of two qualitatively different components, specifically one that is gene-rich and recombinogenic with gene repertoire and order that is still recognizably similar to those in members of other angiosperm families (Vitis, Arabidopsis), and another that is repeat-rich and recombinationally-recalcitrant with relatively few genes that are highly rearranged relative to their homologs in other taxa. This general picture of cotton genome organization is similar to the picture that emerged from comparison of two monocot genomes, rice and sorghum [82, 87].
Curiously, we were able to anchor more contigs on the Vitis genome despite the closer relationship of cotton to Arabidopsis. This difference is attributable in part to differences in anchoring parameters (see Methods), but also reflects the relatively slow evolution of Vitis[89], and highlights the value of the Vitis genome as a botanical model for cross-taxon comparative genomic studies.
The present genome assembly remains somewhat fragmented and may be further improved as more information and new technology emerges. Adding more genetically anchored STS to the BACs, as well as mapping of more BAC-derived sequences will permit anchoring of more contigs to their corresponding chromosomal locations.
Further improvements of the genetic-physical map
While contigs covering ~40% of the genome have been genetically anchored, a higher density of genetic markers may permit anchoring of many more contigs. Some genetically mapped probes hit only singleton BACs and were not incorporated into the physical map in the interest of minimizing false positives. Nearly 1000 probes that hybridized to GR BACs are from sequences that have not yet been genetically mapped, so are not useful in linking the genetic and physical maps. Designing new overgo probes from mapped sequence-tagged sites can be done recursively as more densely populated genetic maps become available. Conversely, new SSR markers can be developed from BES and put onto the genetic map, which would help anchor more contigs and help confirm the position of those already anchored.
Probes targeted at specific regions of interest
Marker density on the physical map reflects efforts to enrich specific genomic regions containing genes of interest for DNA markers. Most prominent are probes aimed at the Li1 (Ligon lintless-1) and Li2 (Ligon lintless-2) genes of cotton. About 300 overgo probes were designed from genetic markers and EST reads that showed relationship to the regions of these genes. This enrichment created "hotspots" where more GR contigs could be aligned to both Arabidopsis and Vitis (Figure 3). In the AT genome, there is an excess of anchored GR contigs near the bottom of chromosome 2, the upper and lower parts of chromosome 3, and the tip of chromosome 5. These four regions were identified in earlier studies [88] to have been produced by two rounds of whole-genome duplication, all belonging to the consensus group β4. Likewise, the regions near the top of VV chromosome 13 and bottom of chromosome 8 anchor a higher than average number of GR contigs.
A closer look at these "hotspots" revealed that the majority of the contigs anchored here contain probes from the Li1 and Li2 regions. There are 114 contigs anchored in the AT regions described above, 94 (82.5%) of which contains Li1 and/or Li2 probes. In 87 out of these 94 cases, the Li probes provided one or more anchor point(s) in the microsynteny detection. In grape, a total of 134 contigs fell into the most densely anchored regions on grape chromosome 6, 8 and 13; 111 (82.8%) of these contigs contain Li1 or Li2 probes of which 92 provided one or more anchor point(s) in microsynteny detection. Compared to the whole-genome average of 23% (970) contigs that contains Li probes, these regions shows a significant enrichment in Li contigs and the ability to align to the AT and VV genomes.
This illustrates the potential use of the contig assembly in cross genome comparisons, and that the power to detect synteny and align contigs across genomes can be greatly increased by targeted enrichment of specific regions for hybridization probes.
The grape genome as a model
Aligning physical map contigs with sequenced genomes has proven informative in several ways [87, 90]. Comparative mapping data and BES alignments to the human genome helped in assigning bovine physical map contigs to their respective chromosomes [90]. The pattern of sorghum physical map contigs along rice chromosomes has given empirical evidence that gene rearrangement is generally deleterious [87]. Cross-species synteny information has also enabled us to make better use of the sequenced genome data on other genomes.
For cotton, Arabidopsis is the most closely-related genome for which a sequence is published as of this writing. The rapid evolution of, and two additional WGD events in, the Arabidopsis lineage may reduce our ability to align these respective genomes. The Vitis genome, on the other hand, evolves relatively slowly [89] and has experienced no WGD events apart from the hexaploidy (γ) event that is likely to be shared by all dicots [86, 91]. The grape genome might prove to be more useful than that of Arabidopsis in comparative genomics across distantly related species.
One disadvantage of using the grape genome as a model for cotton lies in its relative low gene density compared to the Arabidopsis genome. Unlike sorghum and rice, where the euchromatic regions have a similar gene density in both genomes [82, 87], gene density is at least twice as high in Arabidopsis as in Vitis. Gene density across the currently assembled grape pseudomolecules fluctuates from about 20 to 25 genes per 200 kb in higher gene density regions to 10 to 15 genes per 200 kb in lower gene density regions. Similar analysis showed that gene density is uniformly 50 to 60 genes per 200 kb across the Arabidopsis genome, except for the centromeric regions and a few low density points with 30 to 40 genes per 200 kb. This lower gene density in Vitis reduces our ability to anchor cotton contigs, and look for synteny using contig information. Here, we were able to anchor cotton contigs onto most of the gene dense regions of the Vitis genome, but large parts of the low-gene-density chromosomal regions are not covered.
Using the genetic-physical map in gene cloning
Map-based cloning has always been a long and tedious process. The genetic-physical map provides a shortcut by which contigs spanning a target gene region can be readily identified through flanking markers. Markers immediately upstream and downstream of a target gene can be used to identify neighboring anchored contigs, and sequencing of BACs within the contig(s) could provide candidate genes warranting further study (Table 3). In efforts to characterize a gene involved in cotton fiber development, we were able to identify a contig that anchors to the genetic region of interest using this method, and design new genetic markers very close to the gene (unpublished data).
In our efforts to anchor the contigs through probe hybridization, overgo probes were also designed from specific gene sequences (Table 3). Some probes were designed from specific gene families, e.g. COV2311, COV2312, COV1269 and COV1270 were designed from cotton CesA gene. BACs and contigs that contain these sequences were identified, which provide materials for study of these gene families. Probes were also designed to identify contigs that include Arabidopsis trichome gene homologs. Table 3 shows a list of probes and contigs that is directly applicable to the study of specific genes.
The value of the physical map for positional cloning would be further enhanced by anchoring more contigs onto the genetic maps efficiently and accurately. We have provided a framework on which more than 1500 contigs has been aligned. In genomic regions that are of high priority to specific research efforts (positional cloning, etc), many unanchored contigs might be tentatively merged into the anchored contigs, given a lower stringency or higher tolerance for questionable clones, then seeking additional corroborative data such as additional BAC ends, hybridization anchors, or targeted genetic mapping of hybridizing elements. For regions where no contigs have been anchored yet, a simple probing of the library using flanking genetic markers should be able to help build a local genetic-physical map. Contigs upstream and downstream of a target contig can be identified by manually searching for similar contigs at a lower cutoff, and rebuilding the contigs for the region of interest.
Microsynteny information permits one to utilize new ways of developing genetic markers targeted to a region of interest [92] that may be of high value in translating functional information from botanical models to cotton. The contigs aligned to the AT and VV genomes cover about 1/4 of these respective genomes, primarily in regions that are likely to be gene-rich. Earlier research has identified some Arabidopsis genes with well-defined roles in trichome (including root hair) development that approximately correspond to the locations of cotton fiber QTLs. Some of these genes are in regions which showed conserved organization with the GR physical map contigs. e.g. an α-tubulin gene (TUA6) is found in a region spanned by contig1653 and contig3177; the TTG2 gene, which is involved in trichome pattern formation [93], is in a region spanned by contig937; the ACT2 gene, which involves in trichome morphogenesis [94], is in a region spanned by contig908; the GL2 gene is spanned by contig601. These anchorings may provide a good starting point to search for candidate genes and QTLs with similar functions in cotton fiber development, and help elucidate the similarities and differences in trichome formation in different tissues.
Average band number is crucial in agarose based fingerprinting
The use of both agarose based and HICF methods in this physical map assembly gave us the opportunity to directly compare these two methods that have been widely used in genome projects. Using only the agarose based fingerprints, we obtained a large number of small contigs. To test if this is caused by the low band number, we estimated the expected contig number under our conditions. When cutoff = 1e-12, the minimum matching band number required to call an overlap between two clones is 12 in our study. With a tolerance value of 7 and cutoff of 1e-12, the expected contig number in the assembly would be over 9000 if the average band number per clone is 17. In other words, our agarose based contigging yielded the expected result.
The expected contig number drops rapidly with increased average band number. From the Lander-Waterman formula (seem Methods), if the average band number is increased to 20, the expected contig number would be about 5000. With an average band number of 30, one would expect only about 400 contigs. This should be an underestimation because we are not considering physical gaps and under-represented parts of the genome in the BAC libraries, but nevertheless, shows how critical band numbers are to an agarose-based fingerprinting project. BACs with fewer than 8 bands offer too little information to form statistically-supported contigs, even with identical band patterns.
Our success with using HICF in a targeted manner to improve the physical map stems from much higher band numbers. HICF merged contig-end BAC pairs had average agarose band numbers that are not significantly different from the overall band number (18.02 vs. 18.15 in all BACs in contigs). The reason why they failed to join is due to the high percentage of matching bands needed to call an overlap. FPC was unable to call an overlap even if 11 bands were matching (Figure 1C).
Conclusion
The first genetically anchored whole-genome physical map of a cotton species was built through integration of agarose-based fingerprinting and 'high information content fingerprinting' (HICF). Integrating genetically-anchored overgo hybridization probes and BAC end sequences permitted many physical map contigs to be aligned to a consensus cotton genetic map as well as Arabidopsis and Vitis genome sequences. The cotton genome appears to include two qualitatively different components, specifically one that is gene-rich and recombinogenic with gene repertoire and order still recognizably similar to those in members of other angiosperm families, and another that is repeat-rich and recombinationally-recalcitrant with relatively few genes that are highly rearranged relative to those of other angiosperms. While Vitis appears to be a more informative comparator regarding cotton genome organization, translational genomics from Arabidopsis offers singular benefits in identifying the functions of cotton genes. In summary, the physical map is (a) a link that connects genetic map information with physical sequences; (b) a means of validating/directing whole-genome shotgun sequencing assembly; and (c) a tool providing insight into the genome organization of cotton, in advance of a whole-genome sequence.
Methods
BAC library construction
The Gossypium raimondii (GR) BAC library was constructed by D.G.P. according to Peterson et al.[84]. The library consists of 92,160 individually-archived clones and is available through the Plant Genome Mapping Laboratory http://www.plantgenome.uga.edu. To estimate mean insert size and false positive percentage, two clones were selected from each of the library's 240 384-well plates, and minipreps of these clones were digested with Not I and analyzed by pulsed-field gel electrophoresis. Of the 480 digested clones, 448 produced interpretable banding patterns; the remaining 32 were not visible on the gels suggesting that the DNA was lost in the miniprep procedure. Three of the 448 clones appear to be false positives.
Probe design and hybridization
A total of 2828 sequence-tagged site probes were hybridized to the GR library: 357 were overgos designed from Arabidopsis genic sequences (prefixed AOG); 1751 were designed from genetically mapped cotton markers (prefixed COV for cotton overgos, or CM/COAU/PAR for PCR based probes); and 252 from cotton EST sequence reads (prefixed COV). The rest were designed and probed from cotton genes of interest related to multiple projects. Overgo probes [95] were designed and hybridized to the libraries as described [87]. Briefly, source sequences were aligned to all known plant sequences to using BLAST to find conserved domains, and compared to known plant repeats to screen out possible repetitive sequences. The selected sequences were then chopped into 40 bp segments and screened for GC content of between 40% and 60%.
Probes were labeled using P-32 and applied to macroarrays of 18,432 BACs per membrane in a multiplex of 576 probes, using pools of 24 probes per bottle, by rows, columns and diagonals of a 24 × 24 array of probes. Films were manually scored, scores digitized using text-recognition software (ABBYY FINEREADER), and data deconvoluted and stored in the MS Access database system "BACMan".
Fingerprinting
Agarose based fingerprinting methods were adapted from Marra et al.[85]. Plasmids were extracted in batches of 96-well plates and digested using Hin dIII. Fragments were separated on a 121-lane 1% agarose TAE gel, with a size standard every 5 lanes. Band migration distances and molecular weights were digitized using IMAGE [96], before importing into FPC (F ingerp rinted C ontigs) [97, 98].
High information-content fingerprinting (HICF) was adapted from published methods [99]. Plasmids were digested with EcoRI, BamHI, XbaI, XhoI and HhaI. The ends of restriction fragments were differentially labeled using fluorochrome tagged ddNTPs after the first four enzyme cuts, and the last enzyme further reduced fragment size and produced a blunt end. Fingerprints were generated using an ABI3730xl sequencer and size files generated by GeneMapper v4.0 after processing the chromatograms.
Cross-well contaminations and chimeric clones in HICF
Cross-well contaminations seem to be a more severe problem in HICF than in agarose-based FPC assembly. In our first HICF assemblies, we encountered a very large contig containing as many as ~50% of all BACs, depending on the assembly stringency. To overcome this issue, 1166 BACs were excluded from the assembly due to suspiciously high band numbers (possible chimeras) or by a newly implemented function in FPC to identify potential cross-well contaminations. The new assembly show no contigs containing >24 BACs.
Physical map assembly
Agarose-based fingerprints were assembled first by FPC using a cut-off value of 1e-10 and a tolerance value of 8. CpM (contigs plus markers) tables were used to integrate the marker hybridization results: the cut-off value was relaxed to 1e-8, 1e-7 and 1e-6 when two BACs shared one, two and three markers respectively.
After the preliminary assembly, two BACs from each end of the largest 4608 agarose FPC contigs were subjected to HICF. These fingerprints were assembled separately in FPC using a cut-off value of 1e-50 and a tolerance of 3. Overgo hybridization information was not used in HICF assembly. Results from HICF were formatted into a marker file, and fed into the final, integrated assembly in the same manner as probe hybridization results. In this assembly, cutoff was set to 1e-12 and tolerance was set to 7. CpM tables were used in integrating the data. Cut-off values were relaxed to 1e-10, 1e-9 and 1e-8 when two BACs shared one, two and three markers (or HICF contig) respectively.
In each of the three iterations of assembly, the final stringency settings (tolerance and cut-off) were determined by comparing results of different cut-off and tolerance value combinations. For HICF, tolerance values of 2 through 5 and cut-off value of 1e-20 through 1e-50 were tested; for agarose fingerprints, tolerance values of 6 through 9 were and cut-off value of 1e-10 through 1e-12 were tested. Possible cross-well contaminations were identified and rendered as singletons using the built-in function under "search commands" in FPC v 9.3.
Finalizing the assembly
End-to-end auto-merges were done recursively by lowering the cut-off value one step at a time, from 1e-12 through 1e-6. Singletons were also merged into the assembly recursively using the Keyset-to-FPC function in the FPC program. The CB maps for each contig with 2 or more Q clones were recalculated using a higher stringency cutoff value. Q-contigs were thus split up by FPC into smaller contigs and singletons. This was done recursively by raising the cutoff value by 1 level at a time until each one of the splitted contigs contains no more than 1 Q clone. A tarball containing all data (both agarose-based fingerprints and HICF) is available at http://www.plantgenome.uga.edu/pgml_image_data/.
Simulation of contig number change with average band number
FPC uses the Sulston score [96] as a cutoff criterion to call overlaps,  . This is the probability of finding j matching bands in two BACs with n bands each. Expected contig numbers were predicted using the Lander-Waterman formula [100], E(contig#) = Ne-LN/G·(1-T/L), where G is the genome length (genome size/average band size), L is the average band number; N is the number of BACs fingerprinted and T is the number of bands needed to call an overlap. In our study, the gel length is 5000 bands, the genome size is 880 Mb, and the average band size is 4096 bps (for a 6-cutter).
. This is the probability of finding j matching bands in two BACs with n bands each. Expected contig numbers were predicted using the Lander-Waterman formula [100], E(contig#) = Ne-LN/G·(1-T/L), where G is the genome length (genome size/average band size), L is the average band number; N is the number of BACs fingerprinted and T is the number of bands needed to call an overlap. In our study, the gel length is 5000 bands, the genome size is 880 Mb, and the average band size is 4096 bps (for a 6-cutter).
BAC-end sequencing
Two BACs from each end of the largest 2016 contigs were end-sequenced by the Arizona Genome Institute using methods as previously described [101].
Anchoring contigs onto genetic maps
To achieve a maximum number of anchor points, a 13-linkage-group consensus map of cotton, constructed by integration of At, Dt, and D genome genetic maps [14] was used to anchor contigs. Probes that hit only one BAC in a contig were considered possible hybridization artifacts and were not used; probes that hit 30 or more BACs in the GR library were considered repetitive and were also excluded. 482 BACs with 8 or more different probes hybridized to them were excluded as possible contamination artifacts produced in hybridization. Contigs were aligned to the consensus map using the remaining anchor markers.
On average, we had less than one hybridization marker per contig, and the vast majority of contigs had less than three anchor probes. Thus, instead of requiring the contig to have two or more anchor markers from proximal regions on the genetic map to call an anchor, we listed all the contigs anchored by one or more genetic markers alongside the marker's location(s) on the genetic map.
Aligning contigs to whole-genome sequences
BAC-end sequences (BES) and source sequences of overgo probes were used to BLAST against Arabidopsis thaliana and Vitis vinifera genome sequences, using a penalty score of -2 (instead of -3 as the default value) and a e value of 1e-5 in BLASTn. The penalty score was changed to fit the sequence divergence among genomes surveyed, so that longer hits with lower similarity (66.7%) can be retained. Arabidopsis and Vitis genome sequences were downloaded from TAIR ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes/ and Genoscope http://www.genoscope.cns.fr/externe/Download/Projets/Projet_ML/data/8X/assembly/goldenpath/unmasked/, respectively.
Sequences with 10 or more BLAST hits in either genome were considered repetitive and excluded from later analysis. Probe hybridization results used the same filters described for anchoring to genetic maps. BAC contigs were then linked to the genomic sequences through the BLAST data in a MS Access database query. The query results were processed by a Python script aligning the contigs to a genomic region of AT or VV when two or more sequences from the same contig hit a genomic region less than 200 kb (against AT) or 1 Mbp (against VV) apart.
References
Wendel JF, Albert VA: Phylogenetics of the Cotton genus (Gossypium): Character-state weighted parsimony analysis of chloroplast-DNA restriction site data and its systematic and biogeographic implications. Systematic Botany. 1992, 17: 115-143. 10.2307/2419069.
Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF: Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006, 16: 1252-1261. 10.1101/gr.5282906.
Hendrix B, Stewart JM: Estimation of the nuclear DNA content of gossypium species. Ann Bot (Lond). 2005, 95: 789-797. 10.1093/aob/mci078.
Rong J, Abbey C, Bowers JE, Brubaker CL, Chang C, Chee PW, Delmonte TA, Ding X, Garza JJ, Marler BS: A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics. 2004, 166: 389-417. 10.1534/genetics.166.1.389.
Reinisch A, Dong J-M, Brubaker C, Stelly D, Wendel J, Paterson A: A detailed RFLP map of cotton (Gossypium hirsutum × G. barbadense): Chromosome organization and evolution in a disomic polyploid genome. Genetics. 1994, 138: 829-847.
Brubaker CL, Paterson AH, Wendel JF: Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome. 1999, 42: 184-203. 10.1139/gen-42-2-184.
Desai A, Chee PW, Rong J-K, May OL, Paterson AH: Chromosome structural changes in diploid and tetraploid A genomes of Gossypium. Genome. 2006, 49: 10.1139/G05-116.
Wendel JF: New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA. 1989, 86: 4132-4136. 10.1073/pnas.86.11.4132.
Wendel JF, Schnabel A, Seelanan T: Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc Natl Acad Sci USA. 1995, 92: 280-284. 10.1073/pnas.92.1.280.
Xiao J, Wu K, Fang DD, Stelly DM, Yu J, Cantrell RG: New SSR Markers for Use in Cotton (Gossypium spp.) Improvement. The Journal of Cotton Science. 2009, 13: 75-157.
Lacape JM, Nguyen TB, Thibivilliers S, Bojinov B, Courtois B, Cantrell RG, Burr B, Hau B: A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum × Gossypium barbadense backcross population. Genome. 2003, 46: 612-626. 10.1139/g03-050.
Guo WZ, Cai CP, Wang CB, Han ZG, Song XL, Wang K, Niu XW, Wang C, Lu KY, Shi B, Zhang TZ: A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in gossypium. Genetics. 2007, 176: 527-541. 10.1534/genetics.107.070375.
Yu JW, Yu SX, Lu CR, Wang W, Fan SL, Song MZ, Lin ZX, Zhang XL, Zhang JF: High-density linkage map of cultivated allotetraploid cotton based on SSR, TRAP, SRAP and AFLP markers. Journal of Integrative Plant Biology. 2007, 49: 716-724. 10.1111/j.1744-7909.2007.00459.x.
Rong J, Bowers JE, Schulze SR, Waghmare VN, Rogers CJ, Pierce GJ, Zhang H, Estill JC, Paterson AH: Comparative genomics of Gossypium and Arabidopsis: unraveling the consequences of both ancient and recent polyploidy. Genome Res. 2005, 15: 1198-1210. 10.1101/gr.3907305.
Endrizzi J, Ramsay G: Monosomes and telosomes for 18 of the 26 chromosomes of Gossypium hirsutum. Canadian Journal of Genetics. 1979, 21: 531-536.
Saha S, Raska D, Stelly DM: Upland (Gossypium hirsutum L.) × Hawaiian cotton (G. tomentosum Nutt. ex Seem.) F1 hybrid hypoaneuploid chromosome substitution series. Journal of Cotton Science. 2006, 10: 263-272.
Wang K, Song XL, Han ZG, Guo WZ, Yu JZ, Sun J, Pan JJ, Kohel RJ, Zhang TZ: Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theoretical and Applied Genetics. 2006, 113: 73-80. 10.1007/s00122-006-0273-7.
Jiang CX, Wright RJ, El-Zik KM, Paterson AH: Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proceedings of the National Academy of Sciences of the United States of America. 1998, 95: 4419-4424. 10.1073/pnas.95.8.4419.
Draye X, Chee P, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH: Molecular dissection of interspecific variation between Gossypium hirsutum and G-barbadense (cotton) by a backcross-self approach: II. Fiber fineness. Theoretical and Applied Genetics. 2005, 111: 764-771. 10.1007/s00122-005-2061-1.
He DH, Lin ZX, Zhang XL, Zhang YX, Li W, Nie YC, Guo XP: Dissection of genetic variance of fibre quality in advanced generations from an interspecific cross of Gossypium hirsutum and G-barbadense. Plant Breeding. 2008, 127: 286-294. 10.1111/j.1439-0523.2007.01452.x.
Mir RR, Rustgi S, Sharma S, Singh R, Goyal A, Kumar J, Gaur A, Tyagi AK, Khan H, Sinha MK: A preliminary genetic analysis of fibre traits and the use of new genomic SSRs for genetic diversity in jute. Euphytica. 2008, 161: 413-427. 10.1007/s10681-007-9597-x.
Guo YF, McCarty JC, Jenkins JN, Saha S: QTLs for node of first fruiting branch in a cross of an upland cotton, Gossypium hirsutum L., cultivar with primitive accession Texas 701. Euphytica. 2008, 163: 113-122. 10.1007/s10681-007-9613-1.
Qin HD, Guo WZ, Zhang YM, Zhang TZ: QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theoretical and Applied Genetics. 2008, 117: 883-894. 10.1007/s00122-008-0828-x.
Asif M, Mirza JI, Zafar Y: Genetic analysis for fiber quality traits of some cotton genotypes. Pakistan Journal of Botany. 2008, 40: 1209-1215.
Paterson AH, Saranga Y, Menz M, Jiang CX, Wright RJ: QTL analysis of genotype × environment interactions affecting cotton fiber quality. Theoretical and Applied Genetics. 2003, 106: 384-396.
Abdurakhmonov IY, Buriev ZT, Saha S, Pepper AE, Musaev JA, Almatov A, Shermatov SE, Kushanov FN, Mavlonov GT, Reddy UK: Microsatellite markers associated with lint percentage trait in cotton, Gossypium hirsutum. Euphytica. 2007, 156: 141-156. 10.1007/s10681-007-9361-2.
Abdurakhmonov IY, Kohel RJ, Yu JZ, Pepper AE, Abdullaev AA, Kushanov FN, Salakhutdinov IB, Buriev ZT, Saha S, Scheffler BE: Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics. 2008, 92: 478-487. 10.1016/j.ygeno.2008.07.013.
Chee P, Draye X, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH: Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theoretical and Applied Genetics. 2005, 111: 757-763. 10.1007/s00122-005-2063-z.
Chee PW, Draye X, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH: Molecular dissection of phenotypic variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: III. Fiber length. Theoretical and Applied Genetics. 2005, 111: 772-781. 10.1007/s00122-005-2062-0.
Guo WZ, Zhang TZ, Shen XL, Yu JZ, Kohel RJ: Development of SCAR marker linked to a major QTL for high fiber strength and its usage in molecular-marker assisted selection in upland cotton. Crop Science. 2003, 43: 2252-2256. 10.2135/cropsci2003.2252.
He DH, Lin ZX, Zhang XL, Nie YC, Guo XP, Feng CD, Stewart JM: Mapping QTLs of traits contributing to yield and analysis of genetic effects in tetraploid cotton. Euphytica. 2005, 144: 141-149. 10.1007/s10681-005-5297-6.
He DH, Lin ZX, Zhang XL, Nie YC, Guo XP, Zhang YX, Li W: QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × Gossypium barbadense. Euphytica. 2007, 153: 181-197. 10.1007/s10681-006-9254-9.
Kohel RJ, Yu J, Park YH, Lazo GR: Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica. 2001, 121: 163-172. 10.1023/A:1012263413418.
Mei M, Syed NH, Gao W, Thaxton PM, Smith CW, Stelly DM, Chen ZJ: Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theoretical and Applied Genetics. 2004, 108: 280-291. 10.1007/s00122-003-1433-7.
Ren LH, Guo WZ, Zhang TZ: Identification of quantitative trait loci (QTLs) affecting yield and fiber properties in chromosome 16 in cotton using substitution line. Acta Botanica Sinica. 2002, 44: 815-820.
Saha S, Jenkins JN, Wu J, McCarty JC, Stelly DM: Genetic analysis of agronomic and fibre traits using four interspecific chromosome substitution lines in cotton. Plant Breeding. 2008, 127: 612-618. 10.1111/j.1439-0523.2008.01532.x.
Shen XL, Guo WZ, Lu QX, Zhu XF, Yuan YL, Zhang TZ: Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton. Euphytica. 2007, 155: 371-380. 10.1007/s10681-006-9338-6.
Shen XL, Guo WZ, Zhu XF, Yuan YL, Yu JZ, Kohel RJ, Zhang TZ: Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Molecular Breeding. 2005, 15: 169-181. 10.1007/s11032-004-4731-0.
Shen XL, Zhang TZ, Guo WZ, Zhu XF, Zhang XY: Mapping fiber and yield QTLs with main, epistatic, and QTL × environment interaction effects in recombinant inbred lines of upland cotton. Crop Science. 2006, 46: 61-66. 10.2135/cropsci2005.0056.
Ulloa M, Saha S, Jenkins JN, Meredith WR, McCarty JC, Stelly DM: Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum L.) joinmap. Journal of Heredity. 2005, 96: 132-144. 10.1093/jhered/esi020.
Wan Q, Zhang ZS, Hu M, Chen L, Liu DJ, Chen X, Wang W, Zheng J: T-1 locus in cotton is the candidate gene affecting lint percentage, fiber quality and spiny bollworm (Earias spp.) resistance. Euphytica. 2007, 158: 241-247. 10.1007/s10681-007-9446-y.
Wang BH, Guo WZ, Zhu XF, Wu YT, Huang NT, Zhang TZ: QTL mapping of fiber quality in an elite hybrid derived-RIL population of upland cotton. Euphytica. 2006, 152: 367-378. 10.1007/s10681-006-9224-2.
Wang BH, Wu YT, Guo WZ, Zhu XF, Huang NT, Zhang TZ: QTL analysis and epistasis effects dissection of fiber qualities in an elite cotton hybrid grown in second generation. Crop Science. 2007, 47: 1384-1392. 10.2135/cropsci2006.10.0647.
Wu JX, Jenkins JN, McCarty JC, Zhong M, Swindle M: AFLP marker associations with agronomic and fiber traits in cotton. Euphytica. 2007, 153: 153-163. 10.1007/s10681-006-9250-0.
Zhang TZ, Yuan YL, Yu J, Guo WZ, Kohel RJ: Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection. Theoretical and Applied Genetics. 2003, 106: 262-268.
Zhao XQ, Xu JL, Zhao M, Lafitte R, Zhu LH, Fu BY, Gao YM, Li ZK: QTLs affecting morph-physiological traits related to drought tolerance detected in overlapping introgression lines of rice (Oryza sativa L.). Plant Science. 2008, 174: 618-625. 10.1016/j.plantsci.2008.03.009.
Saranga Y, Jiang CX, Wright RJ, Yakir D, Paterson AH: Genetic dissection of cotton physiological responses to arid conditions and their inter-relationships with productivity. Plant Cell and Environment. 2004, 27: 263-277. 10.1111/j.1365-3040.2003.01134.x.
Saranga Y, Menz M, Jiang CX, Wright RJ, Yakir D, Paterson AH: Genomic dissection of genotype × environment interactions conferring adaptation of cotton to arid conditions. Genome Research. 2001, 11: 1988-1995. 10.1101/gr.157201.
Wang HM, Lin ZX, Zhang XL, Chen W, Guo XP, Nie YC, Li YH: Mapping and quantitative trait loci analysis of verticillium wilt resistance genes in cotton. Journal of Integrative Plant Biology. 2008, 50: 174-182. 10.1111/j.1744-7909.2007.00612.x.
Yang C, Guo WZ, Li GY, Gao F, Lin SS, Zhang TZ: QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Science. 2008, 174: 290-298. 10.1016/j.plantsci.2007.11.016.
Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Reddy OUK: Mapping of verticillium wilt resistance genes in cotton. Plant Science. 2005, 168: 1581-1590. 10.1016/j.plantsci.2005.02.008.
Rungis D, Llewellyn D, Dennis ES, Lyon BR: Investigation of the chromosomal location of the bacterial blight resistance gene present in an Australian cotton (Gossypium hirsutum L.) cultivar. Australian Journal of Agricultural Research. 2002, 53: 551-560. 10.1071/AR01121.
Wright R, Thaxton P, El-Zik K, Paterson AH: D-subgenome bias of Xcm resistance genes in tetraploid Gossypium (Cotton) suggests that polyploid formation has created novel avenues for evolution. Genetics. 1998, 149: 1987-1996.
Niu C, Lister HE, Nguyen B, Wheeler TA, Wright RJ: Resistance to Thielaviopsis basicola in the cultivated A genome cotton. Theoretical and Applied Genetics. 2008, 117: 1313-1323. 10.1007/s00122-008-0865-5.
Niu C, Hinchliffe DJ, Cantrell RG, Wang CL, Roberts PA, Zhang JF: Identification of molecular markers associated with root-knot nematode resistance in upland cotton. Crop Science. 2007, 47: 951-960. 10.2135/cropsci2006.07.0499.
Shen XL, Van Becelaere G, Kumar P, Davis RF, May OL, Chee P: QTL mapping for resistance to root-knot nematodes in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theoretical and Applied Genetics. 2006, 113: 1539-1549. 10.1007/s00122-006-0401-4.
Wang C, Ulloa M, Roberts PA: Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton (Gossypium hirsutum L.). Theoretical and Applied Genetics. 2006, 112: 770-777. 10.1007/s00122-005-0183-0.
Wang CL, Roberts PA: Development of AFLP and derived CAPS markers for root-knot nematode resistance in cotton. Euphytica. 2006, 152: 185-196. 10.1007/s10681-006-9197-1.
Ynturi P, Jenkins JN, McCarty JC, Gutierrez OA, Saha S: Association of root-knot nematode resistance genes with simple sequence repeat markers on two chromosomes in cotton. Crop Science. 2006, 46: 2670-2674. 10.2135/cropsci2006.05.0319.
Feng C, Stewart JM, Zhang J: STS markers linked to the Rf1 fertility restorer gene of cotton. Theor Appl Genet. 2005, 110: 237-243. 10.1007/s00122-004-1817-3.
Zhang J, Stewart JM: Identification of molecular markers linked to the fertility restorer genes for CMS-D8 in cotton. Crop Science. 2004, 44: 1209-1217. 10.2135/cropsci2004.1209.
Lan T-H, Cook CG, Paterson AH: Identification of a RAPD marker linked to a male fertility restoration gene in cotton (Gossypium hirsutum L.). J Agric Genom. 1999, 4: 1-5.
Guo WZ, Zhang TZ, Pan JJ, Kohel RJ: Identification of RAPD marker linked with fertility-restoring gene of cytoplasmic male sterile lines in upland cotton. Chinese Science Bulletin. 1998, 43: 52-54. 10.1007/BF02885512.
Wang F, Stewart JM, Zhang J: Molecular markers linked to the Rf(2) fertility restorer gene in cotton. Genome. 2007, 50: 818-824. 10.1139/G07-061.
Desai A, Chee PW, May OL, Paterson AH: Correspondence of trichome mutations in diploid and tetraploid cottons. Journal of Heredity. 2008, 99: 182-186. 10.1093/jhered/esm112.
Lacape JM, Nguyen TB: Mapping quantitative trait loci associated with leaf and stem pubescence in cotton. Journal of Heredity. 2005, 96: 441-444. 10.1093/jhered/esi052.
Wright RJ, Thaxton PM, El-Zik KH, Paterson AH: Molecular mapping of genes affecting pubescence of cotton. Journal of Heredity. 1999, 90: 215-219. 10.1093/jhered/90.1.215.
Ali I, Kausar A, Zafar Y, Asif M, Ashraf M, Riaz S, Zafar S, Wahid A, Maqsood S: Development of Genetic Linkage Map of Leaf Hairiness in Gossypium Hirsutum (Cotton) Using Molecular Markers. Pakistan Journal of Botany. 2009, 41: 1627-1635.
Waghmare VN, Rong J, Rogers CJ, Pierce GJ, Wendel JF, Paterson AH: Genetic mapping of a cross between Gossypium hirsutum (cotton) and the Hawaiian endemic, Gossypium tomentosum. Theor Appl Genet. 2005, 111: 665-676. 10.1007/s00122-005-2032-6.
Jiang C, Wright RJ, Woo SS, DelMonte TA, Paterson AH: QTL analysis of leaf morphology in tetraploid Gossypium (cotton). Theor Appl Genet. 2000, 100: 409-418. 10.1007/s001220050054.
Hao JJ, Yu SX, Dong ZD, Fan SL, Ma QX, Song MZ, Yu JW: Quantitative inheritance of leaf morphological traits in upland cotton. Journal of Agricultural Science. 2008, 146: 561-569.
Song XL, Guo WZ, Han ZG, Zhang TZ: Quantitative trait loci mapping of leaf morphological traits and chlorophyll content in cultivated tetraploid cotton. Journal of Integrative Plant Biology. 2005, 47: 1382-1390. 10.1111/j.1744-7909.2005.00172.x.
Ali I, Ashraf M, Zafar Y, Asif M, Kausar A, Riaz S, Niaz M, Wahid A, Abbas SQ: Development of Genetic Linkage Map of Leaf Red Colour in Cotton (Gossypium Hirsutum) Using DNA Markers. Pakistan Journal of Botany. 2009, 41: 1127-1136.
Sajid Ur R, Malik TA, Ashraf M, Ahsan M: Identification of DNA marker for nectariless trait in cotton using random amplified polymorphic DNA. Pakistan Journal of Botany. 2008, 40: 1711-1719.
Song XL, Zhang TZ: Identification of quantitative trait loci controlling seed physical and nutrient traits in cotton. Seed Science Research. 2007, 17: 243-251. 10.1017/S0960258507834957.
Rong J, Feltus EA, Waghmare VN, Pierce GJ, Chee PW, Draye X, Saranga Y, Wright RJ, Wilkins TA, May OL: Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics. 2007, 176: 2577-2588. 10.1534/genetics.107.074518.
Paterson AH: Sequencing the cotton genomes. World Cotton Research Conference; Lubbock TX. 2007, International Cotton Advisory Committee
Chen ZJ, Scheffler BE, Dennis E: Toward Sequencing cotton (Gossypium) Genomes. Plant Physiology. 2007, 145: 1303-1310. 10.1104/pp.107.107672.
Geever RF, Katterman FRH, Endrizzi JE: DNA hybridization analyses of a Gossypium allotetraploid and two closely related diploid species. Theor Appl Genet. 1989, 77: 553-559. 10.1007/BF00274278.
Walbot V, Dure LS: Developmental Biochemistry of Cotton Seed Embryogenesis and Germination .7. Characterization of Cotton Genome. Journal of Molecular Biology. 1976, 101: 503-536. 10.1016/0022-2836(76)90242-4.
Zhao XP, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, Wendel JF, Paterson AH: Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research. 1998, 8: 479-492.
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A: The Sorghum bicolor genome and the diversification of grasses. Nature. 2009, 457: 551-556. 10.1038/nature07723.
Xu Z, Kohel RJ, Song G, Cho J, Yu J, Yu S, Tomkins J, Yu JZ: An integrated genetic and physical map of homoeologous chromosomes 12 and 26 in Upland cotton (G. hirsutum L.). BMC Genomics. 2008, 9: 108-10.1186/1471-2164-9-108.
Peterson DG, Tomkins JP, Frisch DA, A. WR, Paterson AH: Construction of Plant Bacterial Artificial Chromosome (BAC) Libraries: An Illustrated Guide. 2000, 5:
Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH: High throughput fingerprint analysis of large-insert clones. Genome Res. 1997, 7: 1072-1084.
Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH: Synteny and collinearity in plant genomes. Science. 2008, 320: 486-488. 10.1126/science.1153917.
Bowers JE, Arias MA, Asher R, Avise JA, Ball RT, Brewer GA, Buss RW, Chen AH, Edwards TM, Estill JC: Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci USA. 2005, 102: 13206-13211. 10.1073/pnas.0502365102.
Bowers JE, Chapman BA, Rong J, Paterson AH: Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003, 422: 433-438. 10.1038/nature01521.
Tang HB, Wang XY, Bowers JE, Ming R, Alam M, Paterson AH: Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Research. 2008, 18: 1944-1954. 10.1101/gr.080978.108.
Snelling WM, Chiu R, Schein JE, Hobbs M, Abbey CA, Adelson DL, Aerts J, Bennett GL, Bosdet IE, Boussaha M: A physical map of the bovine genome. Genome Biol. 2007, 8: R165-10.1186/gb-2007-8-8-r165.
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C: The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007, 449: 463-467. 10.1038/nature06148.
Feltus FA, Singh HP, Lohithaswa HC, Schulze SR, Silva TD, Paterson AH: A comparative genomics strategy for targeted discovery of single-nucleotide polymorphisms and conserved-noncoding sequences in orphan crops. Plant Physiol. 2006, 140: 1183-1191. 10.1104/pp.105.074203.
Ishida T, Hattori S, Okada K, Wada T: Role of TTG2 in genetic network of epidermal cell differentiation in Arabidopsis. Plant and Cell Physiology. 2007, 48: S82-S82.
Nishimura T, Yokota E, Wada T, Shimmen T, Okada K: A semi-dominant mutation in the ACT2 gene affects the root hair development in Arabidopsis. Plant and Cell Physiology. 2003, 44: S205-S205.
Cai WW, Reneker J, Chow CW, Vaishnav M, Bradley A: An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics. 1998, 54: 387-397. 10.1006/geno.1998.5620.
Sulston J, Mallett F, Durbin R, Horsnell T: Image analysis of restriction enzyme fingerprint autoradiograms. Comput Appl Biosci. 1989, 5: 101-106.
Soderlund C, Humphray S, Dunham A, French L: Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000, 10: 1772-1787. 10.1101/gr.GR-1375R.
Soderlund C, Longden I, Mott R: FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997, 13: 523-535.
Luo MC, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OD, Dvorak J: High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics. 2003, 82: 378-389. 10.1016/S0888-7543(03)00128-9.
Lander ES, Waterman MS: Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988, 2: 231-239. 10.1016/0888-7543(88)90007-9.
Ammiraju JSS, Luo MZ, Goicoechea JL, Wang WM, Kudrna D, Mueller C, Talag J, Kim H, Sisneros NB, Blackmon B: The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Research. 2006, 16: 140-147. 10.1101/gr.3766306.
Acknowledgements
We thank numerous members of the Paterson lab for additional feedback and contributions. We would also like to thank the Dvorak lab of UC-Davis, especially Dr. Jan Dvorak and Dr. Mingcheng Luo, for their support and training in HICF techniques. The authors appreciate financial support for this research from the National Science Foundation (DBI 98-72630, DBI 02-11700 to JFW, AHP, RAW; DBI 02-08311 to AHP), Cotton, Inc (AHP), USDA ARS-58-6402-7-241 (DGP), and the Pakistan-US Science and Technology Cooperation Program (AHP, MR).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AHP, RAW and JFW conceived and designed the experiments; DGP and CL constructed and prepared the BAC library; GJP, ROC and LL generated BAC fingerprints; JCE, JEB, LKR, CK, SK, LL, CG, NT and MR designed and performed the overgo hybridizations; AHC, EE, KLH, JI, JO, EU and SK scored the hybridization results; MB, KC, WG, DK, MW, YY and AZ performed BAC-end sequencing; LL, JEB and HT analyzed the data. Reagents/materials/analysis tools were provided by AHP, JCE, HT, XW and RAW; LL and AHP wrote the manuscript. All authors read and approve the final manuscript.
Electronic supplementary material
12864_2010_2989_MOESM1_ESM.PDF
Additional file 1: The distribution of contigs sizes of the integrated assembly. Contig sizes were measured by the number of BACs contained in one contig. The majority of contigs contain between 3 and 25 BACs. (PDF 229 KB)
12864_2010_2989_MOESM2_ESM.XLS
Additional file 2: Sources of the 46 highly repetitive probes. Probes with >100 BAC hits in the GR library and the locus from which the probes were derived. (XLS 24 KB)
12864_2010_2989_MOESM3_ESM.PDF
Additional file 3: A consensus genetic-physical map of the cotton genome. The position of the 1585 genetically anchored physical map contigs on the consensus genetic map integrating the At, Dt and D genome genetic maps. (PDF 532 KB)
12864_2010_2989_MOESM4_ESM.PDF
Additional file 4: GR chloroplast contig. Contig11556 is identified as a chloroplast contig, with BAC-end sequences and an overgo probe aligned to the GH chloroplast sequence. (PDF 393 KB)
12864_2010_2989_MOESM5_ESM.PDF
Additional file 5: Gene ontology analysis result of the G. raimondii sequences. GO classification results generated from 13662 BAC-end sequences and 13661 random shotgun sequences, using Blast2Go at an ontology level of 2. (Details refer to additional file 6). (PDF 2 MB)
12864_2010_2989_MOESM6_ESM.DOC
Additional file 6: G. raimondii preliminary gene ontology classification results. GO classification results generated from 13662 BAC-end sequences and 13661 random shotgun sequences, using Blast2Go at an ontology level of 2. (DOC 36 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lin, L., Pierce, G.J., Bowers, J.E. et al. A draft physical map of a D-genome cotton species (Gossypium raimondii). BMC Genomics 11, 395 (2010). https://doi.org/10.1186/1471-2164-11-395
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-11-395