Abstract
Background
A close association between Sst I polymorphism in the 3' untranslated region of the apolipoproteinC3 (APOC3 ) gene and levels of plasma triglycerides (TG) had been reported by different investigators. Hypertriglyceridemia(HTG) is a known risk factor for coronary artery disease (CAD) in the context of Asian Indians. We conducted a study on the relationship between APOC3 SstI polymorphism (S1S1, S1S2 and S2S2 genotypes) and plasma TG levels in a group of 139 male healthy volunteers from Northern India.
Methods
DNA samples were analyzed by polymerase chain reaction (PCR) followed by SstI digestion. Digested PCR products were run on 3% agarose gel and visualized by ethidium bromide staining.
Results
Rare S2 allele was highly prevalent in our study population (0.313) as compared to the Caucasians (0.00–0.11). The genotypic distribution was in agreement with Hardy-Weinberg equilibrium . S2 allele was almost two times more prevalent in the HTG group (N = 34) as compared to NTG group (N = 105) (p = 0.001). Multiple logistic regression revealed S1S2 individuals had age-adjusted odds ratio of 2.43 (95%CI = 0.99–6.01, p = 0.054) and S2S2 had 9.9 (95%CI = 2.66–37.29, p = 0.0006) for developing HTG in comparison to S1S1 genotype.
Conclusions
Our study shows a significant association between rare S2 allele and HTG in Asian Indians.
Similar content being viewed by others
Background
Apolipoprotein CIII (apoCIII protein; APOC3 gene) is a 79 amino acids long glycoprotein that is synthesized predominantly in the liver and to a lesser degree in the intestine [1]. It is present on very low density lipoproteins (VLDLs) and chylomicron remnants; and to some extent on high density lipoproteins (HDLs) [1]. Although the precise function of apoCIII is not clearly understood, several lines of evidence suggest its involvement in the regulation of triglyceride (TG) levels. In vitro , apoCIII inhibits lipoprotein lipase (LPL), a rate-limiting enzyme for TG hydrolysis, resulting in the delayed catabolism of TG-rich particles [2]. Furthermore, it also decreases apoE-mediated remnant removal by displacement of apoE from the VLDL particles in vivo [3, 4]. Additional copies of human APOC3 gene in transgenic mice were associated with hypertriglyceridemia (HTG) [5], whereas the absence of the gene in knock out mice leads to reduced TG [6].
APOC3 gene has been mapped on the long arm of chromosome11, closely linked to the APOA4 and APOA1 genes [7]. A transversion from C to G in the 3' untranslated region (3'UTR) of exon 4 in the APOC3 gene results in an SstI polymorphism. Several studies have suggested a close association between rare S2 allele of SstI polymorphism and elevated TG levels [8–24] and apoCIII levels [22–25]. However, few other studies have shown contradictory results [26–31].
We have investigated the association of APOC3 SstI polymorphism with TG levels in a group of healthy volunteers from Northern India; considering the high prevalence of CAD in Asian Indians, HTG as one of the underlining risk factors in the progression of coronary atherosclerosis [32] and no information available on Asian Indians in this context.
Results
The characteristics of the NTG and HTG groups are shown in Table 1. There was no significant difference in the mean age between the two groups (p = 0.748). TC (p = 0.0001) and TG levels (p < 0.0001) were significantly higher in the HTG group as compared to the NTG group. There was no significant difference in LDL, HDL and LDL/HDL ratio.
The genotypic and allelic distribution of APOC3 polymorphism in the study population is shown in Table 2. No significant deviation from Hardy-Weinberg equilibrium was observed in the study population. The frequency of the S2 allele in the study population was 0.313 (Table 2). The 95% CI of the allele frequency is also presented in Table 2.
To determine the association of APOC3 SstI polymorphism with TG levels, the study population was divided into NTG and HTG groups. Out of 139 individuals, 34 subjects (24.46%) were in HTG group i.e. TG levels beyond 1.921 mmol/L. Table 3 summarize the distribution of various genotypes and alleles of APOC3 polymorphism between the NTG and HTG groups. The 95% CI of the allele frequencies is also presented in Table 3. No significant difference was observed between the expected and observed genotype frequencies of the two groups (NTG: Chi square = 0.9852, df = 1, p = 0.3209 and HTG: Chi square = 0.000, df = 1 and p = 0.9960; in Hardy-Weinberg equilibrium ). There was a significant difference in the genotypic distribution between the NTG and HTG groups as shown in Table 3 (Chi square = 14.019, df = 2, p = 0.001). S1S1 genotype was more frequent in the NTG group as compared to the HTG group (53.3% vs. 26.5%). Conversely, S2S2 genotype was almost five times more prevalent in the HTG group as compared to the NTG group (23.5% vs. 4.8%). However, not much difference was observed in the S1S2 frequency between the two groups (HTG: 50% vs. NTG: 41.9%). In totality, 73.5% of HTG group (25/34) were S2 carriers out of which 32% individuals (i.e. 8/25) were S2 homozygotes. Comparatively, NTG group had 46.7%(49/105) S2 carriers, of which only 10.2%(5/49) were S2 homozygotes. Consequently, S2 allele was almost two times more frequent in the HTG group compared to the NTG group (p = 0.001) as shown in Table 3.
Logistic analysis revealed a significant association of APOC3 S2 allele with hypertriglyceridemia (Table 3). The crude Odds ratio (OR) for S1S2 genotype (in comparison to S1S1 genotype) was found to be 2.4 (95%CI: 0.98–5.90), which was towards significance (p= 0.056) and did not change much after adjusting for age (OR= 2.43, 95% CI = 0.99–6.01, p= 0.054). The crude OR for S2S2 to develop HTG (in comparison to S1S1) was found to be 9.95 (95% CI: 2.66–37.29, p = 0.0006), which was highly significant and remained significant even after adjusting for age (OR = 9.90, 95%CI = 2.64–37.12, p = 0.0007). In totality, S2 carriers had a crude OR of 3.17 to develop HTG (95%CI = 1.35–7.45, p = 0.008) and 3.22 (95% CI = 1.37–7.60, p= 0.007) after age adjustment.
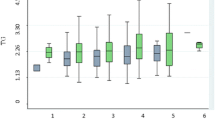
The intergenotypic variations in lipid profile in the HTG, NTG and total subjects are shown in Table 4. TG was significantly different among various genotypes in the HTG (p = 0.015, log TG: p = 0.015), the total subjects (p < 0.0001) and insignificant in the NTG group (p = 0.114, logTG: p = 0.137). In particular, the S2S2 individuals were associated with highest concentration of TG followed by S1S2 and then by S1S1 in HTG group & total study population. No significant differences were observed in TC, LDL, HDL and LDL/HDL ratio in any of the study group (Table 4).
Discussion
ApoCIII provides a strong negative charge on the surface of lipoproteins preventing nonspecific interactions with cell surfaces [33] and perhaps with other lipoproteins. This may serve the function of reducing futile cycles in TG transport by preserving the particles for high affinity interactions such as with lipoprotein lipase or specific cell surface receptors e.g., such as those binding to apoE or apoB. Plasma concentrations of apoC-III in human populations correlate well with TG levels [16, 34]. In vivo apoCIII modulates the postprandial management of the TG [6] and inhibits the hepatic uptake of VLDL remnants [35]. The genetically determined deficiency of apoCIII in humans has been shown to increase the rate of TG clearance from plasma by 6- to 7-fold [36]. A similar enhancement of TG clearance was observed in mice made apoCIII deficient by gene knockout experiments [6]. Overexpression of apoCIII produces hypertriglyceridemia in transgenic mouse models via inhibition of clearance of TG-rich particles [4]. It is now clear that normal physiological systems responsible for TG transport are partially determined by the plasma content of apoCIII. Although it is not clear how this protein contributes to the familial hypertriglyceridemic syndromes, recent studies have found that two classes of drugs that are effective in lowering plasma TG in these patients act through suppression of APOC3 gene transcription in rodents [37, 38].
The human APOC3 gene expression is controlled by positive and negative elements that are spread through out the APOA1-C3-A4 gene cluster on the long arm of chromosome 11 [39]. Various restriction fragment length polymorphisms in and around the human APOC3 gene have been associated with hypertriglyceridemia in several distinct populations [40]. The present study on SstI polymorphism was carried out on a random sample of 139 individuals inhabiting plains of Northern part of India. The genotypic distribution was in good agreement with Hardy-Weinberg equilibrium . Our study revealed a higher frequency of rare S2 allele (0.313) than observed for most of the Caucasians (0.00–0.11) [8–13, 18, 21, 26] but within the same range as reported for different non Caucasian populations (0.15–0.39) [14–17, 20, 28–31]. Earlier Paul et al reported the distribution of APOC3 SstI polymorphism in a much smaller sample of immigrant Asian Indians at U.K. (S2 = 0.19, N = 24) [41]. We attempted at elucidating the association of APOC3 SstI polymorphism with TG levels. Individuals having TG levels up to 1.921 mmol/L were grouped in NTG and more than 1.921 mmol/L were grouped in HTG group. S2S2 individuals had the highest levels of TG followed by S1S2 and S1S1 in HTG and total study population. Significantly higher frequency of S2 allele in the HTG group as compared to the NTG group suggests a strong association of the S2 allele with higher levels of TG. Such an association of S2 allele with higher levels of TG has been reported in studies carried out on Caucasians [8–13, 18, 21–24], Chinese [14], Mayans [15], Japanese [16], Koreans [17], South Africans [19] and Arabs [20]. The biochemical basis for the association of S2 allele with hypertriglyceridemia has yet to be established. Dallinga-Thie et al [22, 23] and Shoulders et al [24, 25] reported an association between levels of apoCIII and S2 allele. The SstI polymorphism is located in the 3' untranslated region of APOC3 gene. Therefore, it is more likely that S2 allele is not etiological but in linkage disequilibrium with other causative mutation hitherto unknown in APOC3 or nearby gene involved in determining the TG levels. It has been suggested that certain haplotypes generated from SstI polymorphism and promoter polymorphism of APOC3 gene may protect or predispose to hypertriglyceridemia [17]. In addition, SstI polymorphism may also influence mRNA stability [17]. Few of the studies carried out on Caucasians [26, 27], Taiwanese [28], Japanese [29, 30] and Arabs [31] did not find any significant association between SstI polymorphism and HTG. It has been speculated that the linkage disequilibrium between this polymorphic site and the causative mutation is weakened or absent in some populations [24].
Ours is the first study on Asian Indians to report a strong association of APOC3 S2 allele with hypertriglyceridemia in Indians. The logistic analysis revealed that individuals carrying S2 allele were 3.2 times more prone to develop hypertriglyceridemia as compared to S1S1. Thus, S2 allele may serve as a significant risk marker for susceptibility to hypertriglyceridemia. This is an important finding as Asian Indians are highly sensitive to the adverse effects of hypertriglyceridemia [32], the risk of which is likely to increase manifold with growing shift towards affluent lifestyle and sedentary habits in larger fraction of our population.
Conclusion
We found a high prevalence of rare S2 allele of APOC3 gene in HTG individuals. Further, this allele was more frequent in our study population as compared to Caucasians. Since HTG is considered as a risk factor for CAD in Asian Indians, there is an urgent need to evaluate the association of APOC3 SstI polymorphism with the risk of developing coronary artery disease in Asian Indians.
Materials and Methods
One hundred and thirty nine healthy male volunteers from plains of northern part of India (mean age: 52.32 ± 11.02 years) were enrolled in the study. The subjects were scrutinized on the basis of standard questionnaire. They shared fairly common socio-cultural background and comparable dietary habits. All underwent routine biochemical tests (hemoglobin, urea & sugar) and blood pressure measurement. Subjects having angina or any history of myocardial infarction were excluded from the study. The research was undertaken with the approval of ethical committee set by All India Institute of Medical Sciences, New Delhi and its guidelines were observed.
Venous blood was collected from each individual after at least 12 hours of fasting. Lipid profile was monitored using enzymatic kits (Randox laboratories limited, UK).
The study subjects were classified into normotriglyceridemic group (NTG: N = 105, TG < = 1.921 mmol/L) and hypertriglyceridemic group (HTG: N = 34, TG>1.921 mmol/L) based on the normal range of the TG calibrated at AIIMS, which is 0.791–1.921 mmol/L. The calibration was done on data obtained from huge number of serum samples. All the chemicals used in the study were procured from Sigma Chemical Co., USA, if not specified.
DNA was extracted from blood by salting out method [42]. 100–500 ng of DNA was amplified in a thermocycler (PTC-100, MJ Research Inc., USA) using 1 unit of Taq DNA Polymerase (Life Technologies Inc., USA) in a 25 ul reaction mixture containing 10 picomole forward primer: 5'-CAT GGT TGC CTA CAG GAG TTC-3'and reverse primer: 3'-TGA CCT TCC GCA CAA AGC TGT-5' (MWG Biotech GmbH, Germany)[8]. The PCR mixture also contained 10% DMSO and 5 mM dNTPs (Life Technologies Inc., USA). DNA was initially denatured for 5 minutes at 95°C, annealed at 58°C for 3 minutes and at 72°C for 5 minutes. The cycling conditions were set to heat the samples at 95°C for 50 seconds, at 58°C for 45 seconds and at 72°C for 1 minute. The cycle was repeated 40 times followed by final extension at 72°C for 10 minutes.
Nine microlitre of the PCR product was digested at 37°C overnight with 10 units of SacI restriction enzyme (New England Biolabs Inc., USA) in the presence of 1 ul of 10X buffer provided with the restriction enzyme. The digested PCR product was resolved on a 3% agarose gel using 1XTBE buffer (89 mM Tris Borate, 2 mM EDTA, pH8.3) at 80 V for at least 1 hour and visualized by ethidium bromide staining on a U. V. transilluminator. The presence of the SstI site yields two fragments of 225 bp and 371 bp, while in the absence of the site one fragment of 596 bp is observed. The wild type allele lacking the restriction site is called as S1 and the allele containing the SstI restriction site is designated as S2 allele.
Allelic frequencies were estimated by gene-counting method. The sample-size dependent standard error of alleles was calculated in terms of 95% confidence interval (CI) of the estimates. Chi-square goodness-of-fit was used to verify the agreement of the observed genotype frequencies with those expected ones (Hardy-Weinberg equilibrium ) in various study groups. Chi-square test was applied to compare genotypic frequencies between the two groups. Contingency table approach (Fisher's RxC test) was used to determine if there is significant differences in allele frequencies among the group of individuals. The biochemical characteristics of the individuals in various genotypic groups were expressed in terms of mean ± standard deviation (S.D.) and were compared using analysis of variance (Annova). Triglycerides values were also log transformed because the test of homogeneity of variance was found to be significant. Logistic analysis with enter method was performed. S1S1 genotype was taken as the reference and the odds ratio with 95% confidence interval was calculated for S1S2 and S2S2 genotypes individually and taken together. Hypertriglyceridemia was entered as dependent variable with NTG = 0 and HTG = 1. All the statistical analysis was preformed using SPSS (Statistical Package for Social Sciences) for windows (version 7.5.10, SPSS Inc., Chicago). Statistical significance was set at p < 0.05.
References
Breslow JL: Familial disorders of high density lipoprotein metabolism. In : The metabolic basis of inherited disease (Edited by: Scriver CR, Beaudet AL, Sly WS, and Valle D). New York, McGraw–Hill 1995, 1251–1266.
Wang CS, McConathy WJ, Kloer HU, Alaupovic P: Modulation of lipoprotein lipase activity by apolipoproteins. Effect of apolipoprotein C–III. J Clin Investig 1985, 75: 384–390.
Aalto–Setala K, Weinstock PH, Bisgaier CL, Wu L, Smith JD, Breslow JL: Further characterization of the metabolic properties of triglyceride–rich lipoproteins from human and mouse apoC–III transgenic mice. J Lipid Res 1996, 37: 1802–1811.
Aalto–Setala K, Fisher EA, Chen X, Chajek–Shaul T, Hayek T, Zechner R, Walsh A, Ramakrishnan R, Ginsberg HN, Breslow JL: Mechanism of hypertriglyceridemia in human apolipoprotein (apo) CIII transgenic mice. Diminished very low density lipoprotein fractional catabolic rate associated with increased apo CIII and reduced apo E on the particles. J Clin Investig 1992, 90: 1889–1900.
Ito Y, Azrolan N, Connell A, Walsh A, Breslow JL: Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science 1990, 249: 790–793.
Maeda N, Li H, Lee D, Oliver P, Quarfordt SH, Osada J: Targeted disruption of the apolipoprotein C–III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J Biol Chem 1994, 269: 23610–23616.
Karanthansis SK: Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII and AIV genes. Proc Natl Acad Sci USA 1985, 82: 6374–6378.
Paul–Hayase H, Rosseneu M, Robinson D, van Bervliet JP, Deslypere JP, Humphries SE: Polymorphisms in the apolipoprotein (apo) AI–CIII–AIV gene cluster: detection of genetic variation determining plasma apo AI, apo CIII and apo AIV concentrations. Hum Genet 1992, 88: 439–446.
Rees A, Stocks J, Sharpe CR, Vella MA, Shoulders CC, Katz J, Jowett NI, Barelle FE, Galton DJ: Deoxyribonucleic acid polymorphism in the apolipoprotein A–1–C–III gene cluster. J Clin Invest 1985, 76: 1090–1095.
Stocks J, Paul–Hayase H, Galton DJ: Haplotypes identified by DNA restriction–fragment–length polymorphisms in the A–I C–III A–IV gene region and hypertriglyceridemia. Am J Hum Genet 1987, 41: 106–118.
Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL: An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 3'untranslated region polymorphisms. Proc Natl Acad Sci USA 1993, 90: 4562–4566.
Tybjaerg–Hansen A, Nordestgaard BG, Gerdes LU, Faergeman O, Humphries SE: Genetic markers in the apoAI–CIII–AIV gene cluster for combined hyperlipidemia, hypertriglyceridemia, and predisposition to atherosclerosis. Atherosclerosis 1993, 100: 157–169.
Hoffer MJV, Sijbrands EJG, de Man FHAF, Havekes LM, Smelt AHM, Frants R: Increased risk for endogenous hypertriglyceridaemia is associated with an apolipoprotein C3 haplotype specified by the SstI polymorphism. Eur J Clin Invest 1998, 28: 807–812.
Ko Y–L, Ko Y–S, Wu S–M, Teng M–S, Chen F–R, Hsu T–S, Chiang C–W, Lee Y–S: Interaction between obesity and genetic polymorphisms in the apolipoprotein CIII gene and lipoprotein lipase gene on the risk of hypertriglyceridemia in Chinese. Hum Genet 1997, 100: 327–333.
Ahn YI, Valdez R, Reddy AP, Cole SA, Weiss KM, Ferrell RE: DNA polymorphisms of the apolipoprotein AI/CIII/AIV gene cluster influences plasma cholesterol and triglyceride levels in the Mayans of the Yucatan peninsula, Mexico. Hum Hered 1991, 41: 281–289.
Zeng Q, Dammerman M, Takada Y, Matsunaga A, Breslow JL, Sasaki J: An apolipoprotein CIII marker associated with hypertriglyceridemia in Caucasians also confers increased risk in a west Japanese population. Hum Genet 1995, 95: 371–375.
Hong SH, Park WH, Lee CC, Song JH, Kim JQ: Association between genetic variations of apo AI–CIII–AIV cluster gene and hypertriglyceridemic subjects. Clin Chem 1997, 43: 13–17.
Rees A, Shoulder CC, Stocks J, Galton DJ, Baralle FE: DNA polymorphisms adjacent to human A–I gene: relation to hypertriglyceridaemia. Lancet 1983, 1: 444–446.
Henderson HE, Landen SV, Michie J, Berger GMB: Association of DNA polymorphism in the apolipopoprotein CIII gene with diverse hyperlipidaemic phenotypes. Hum Genet 1987, 75: 62–65.
Tas S: Strong association of a single nucleotide substitution in the 3'–untranslated region of the apolipoprotein C–III gene with common hypertriglyceridemia in Arabs. Clin Chem 1989, 35: 256–259.
Aalto–Setala K, Kontula K, Sane T, Nieminen M, Nikkila E: DNA polymorphisms of apolipoprotein A–I/C–III and insulin genes in familial hypertriglyceridemia and coronary heart disease. Atherosclerosis 1987, 66: 145–152.
Dallinga–Thie GM, Bu XD, van Linde–Sibenius Trip M, Rotter JI, Lusis AJ, de Bruin TWA: Apolipoprotein A–I/C–III/A–IVgene cluster in familial combined hyperlipidemia: effects on LDL–cholesterol and apolipoproteins B and C–III. J Lipid Res 1996, 37: 136–147.
Dallinga–Thie GM, van Linde–Sibenius Trip M, Rotter JI, Cantor RM, Bu X–D, Lusis AJ, de Bruin TWA: Complex genetic contribution of the Apo AI–CIII–AIV gene cluster to familial combined hyperlipidemia. Identification of different susceptibility haplotypes. J Clin Investig 1997, 99: 953–961.
Shoulders CC, Grantham TT, North JD, Gaspardone A, Tomai F, de Fazio A, Versaci F, Gioffre PA, Cox NJ: Hypertriglyceridemia and the apolipoprotein CIII gene locus: lack of association with the variant insulin response element in Italian school children. Hum Genet 1996, 98: 557–566.
Shoulders CC, Harry PJ, Lagrost L, White SE, Shah NF, North JD, Gilligan M, Gambert P, Ball MJ: Variation at the apo AI/CIII/AIVgene complex is associated with elevated plasma levels of apo CIII. Atherosclerosis 1991, 87: 239–247.
Price WH, Morris SW, Burgon R, Donald PM, Kitchin AH: ApolipoproteinCIII polymorphism and coronary heart disease. Lancet 1986, 2: 1041.
Marcil M, Boucher B, Gagne E, Davignon J, Hayden M, Jenest J: Lack of association of the apolipoprotein A–I–C–III–A–IV gene XmnI and SstI polymorphisms and of the lipoprotein lipase gene mutations in familial combined hyperlipoproteinemia in French Canadian subjects. J Lipid Res 1996, 37: 309–319.
Wu JH, Kao J–T, Wen M–S, Lo S–K: DNA polymorphisms at the apolipoprotein A1–CIII loci in Taiwanese: correlation of plasma APOCIII with triglyceride level and body mass index. J Formos Med Assoc 2000, 99: 367–374.
Rees A, Stocks J, Paul–Hayase H, Ohuchi Y, Galton D: Haplotypes identified by DNA polymorphisms at the apolipoprotein A–I and C–III loci and hypertriglyceridemia: a study in a Japanese population. Hum Genet 1986, 72: 168–171.
Bai H, Saku K, Liu R, Imamura M, Arakawa K: Association between coronary heart disease and the apolipoprotein A–I/C–III/A–IV complex in a Japanese population. Hum Genet 1995, 95: 102–104.
Johansen K, Dunn B, Tan JCY, Kwassi AAA, Skootnicki A, Skootnicki M: Coronary artery disease and apolipoprotein AI–CIII gene polymorphism: a study of Saudi Arabians. Clin Genet 1991, 39: 1–5.
Enas EA, Mehta JL: Malignant coronary artery disease in young Asian Indians. Thoughts on pathogenesis, prevention and therapy. New Engl J Med 1995, 333: 1301–1307.
Shelburne F, Hanks J, Meyers W, Quarfordt S: Effect of apoproteins on hepatic uptake of triglyceride emulsion in the rat. J Clin Invest 1980, 65: 652–658.
Schonfeld G, George PK, Miller J, Reilly P, Witztum J: Apolipoprotein CII and CIII levels in hyperlipoproteinemia. Metab clin exp 1979, 28: 1001–1010.
Windler E, Havel RJ: Inhibitory effects of C apolipoproteins from rats and humans on the uptake of triglyceride–rich lipoproteins and their remnants by the perfused rat liver. J Lipid Res 1985, 26: 556–565.
Ginsberg HN, Le NA, Goldberg IJ, Gibson JC, Rubinstein A, Wang–Iverson P, Norum R, Brown WV: Apolipoprotein B metabolism in subjects with deficiency of Apolipoprotein CIII and A. J Clin Invest 1986, 78: 1287–1295.
Bar–Tana J, Frenkel B, Bishara–Shieban J: The effect of b, b9–tetramethylhexadecanedioic acid (MEDICA 16) on plasma very low density lipoprotein metabolism in rats: role of Apolipoprotein CIII. Biochem J 1994, 298: 409–414.
Bar–Tana J, Rose–Kahn G, Frenkel B, Shafer Z, Fainaru M: Hypolipidemic effect of b, b–methyl–substituted hexadecanedioic acid (MEDICA 16) in normal and nephrotic rats. J Lipid Res 1988, 29: 431–441.
Jong MC, Hofker MH, Havekes LM: Role of ApoCs in Lipoprotein Metabolism Functional Differences Between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol 1999, 19: 472–484.
Surguchov AP, Page GP, Smith L, Wolfgang P, Boerwinkle E: A Polymorphic markers in Apolipoprotein C–III gene flanking regions and hypertriglyceridemia. Arterioscler Thromb Vasc Biol 1996, 16: 941–947.
Paul–Hayase H, Galton D, Stocks J: DNA polymorphic patterns and haplotype arrangements of apoA–1, apoC–III, apoA–IV gene cluster in different ethnic groups. Hum Genet 1987, 75: 264–268.
Miller SA, Dykes DD, Polesky HF: A salting out procedure for extracting DNA from human nucleated cells. Nucleic acid Res 1988, 16: 1215.
Acknowledgement
The financial assistance provided by Jawaharlal Nehru Memorial trust, New Delhi, Lady Tata Memorial Trust, Bombay, India and the Volkswagen foundation, Germany is duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Chhabra, S., Narang, R., Krishnan, L. et al. Apolipoprotein C3 SstI polymorphism and triglyceride levels in Asian Indians. BMC Genet 3, 9 (2002). https://doi.org/10.1186/1471-2156-3-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-3-9