Abstract
Background
The discovery of TCF7L2 as a global type 2 diabetes (T2D) gene hassparked investigations to explore the clinical utility of its variants forguiding the development of new diagnostic and therapeutic strategies.However, interpreting the resulting associations into function still remainsunclear. Canonical Wnt signaling regulates β-catenin and its bindingwith TCF7L2, which in turn is critical for the production of glucagon-likepeptide-1 (GLP-1). This study examines the role of a novel frame-shiftinsertion discovered in a conserved region of WNT16a, and it isproposed that this mutation affects T2D susceptibility in conjunction withgene variants in TCF7L2.
Results
Our results predicted that the insertion would convert the upstream openreading frame in the Wnt16a mRNA to an alternative, in-frame translationinitiation site, resulting in the prevention of nonsense-mediated decay,leading to a consequent stabilization of the mutated WNT16a message. Toexamine the role of Wnt16a in the Wnt signaling pathway, DNA and serumsamples from 2,034 individuals (48% with T2D) from the Sikh Diabetes Studywere used in this investigation. Prevalence of Wnt16a insertion did notdiffer among T2D cases (33%) and controls (32%). However, there was a 3.2fold increase in Wnt16a mRNA levels in pancreatic tissues from the insertioncarriers and a significant increase (70%, p < 0.0001) in luciferaseactivity in the constructs carrying the insertion. The expression of TCF7L2mRNA in pancreas was also elevated (~23-fold) among the insertion carriers(p=0.003).
Conclusions
Our results suggest synergistic effects of WNT16a insertion and theat-risk ‘T’ allele of TCF7L2 (rs7903146) for elevating theexpression of TCF7L2 in human pancreas which may affect theregulation of downstream target genes involved in the development of T2Dthrough Wnt/β-catenin/TCF7L2 signaling pathway. However, furtherstudies would be needed to mechanistically link the two definitively.
Similar content being viewed by others
Background
Transcription factor 7-like 2 (TCF7L2) has been strongly linked to type 2 diabetes(T2D) susceptibility, with an elevated genetic predisposition accounting for 20% ofT2D cases [1]. The association of common intronic variants in the TCF7L2 genewith the increased susceptibility for T2D has been extensively documented in majorethnic groups of the world by several different investigators [2]. Meta-analysis of the published studies estimated the odds ratio (OR) of1.46 (p=5.4x10-140) [3]. TCF7L2 polymorphisms were also significantly linked to diabetesrisk in our own studies in Asian Indian Sikhs [4, 5]. Indeed, our recent Sikh genome-wide association study (GWAS) andmeta-analyses in Sikhs (n=7,329/3,354 cases) and South Asians (n=47,303/19, 482cases) showed a robust association of TCF7L2 (rs7903146), with OR 1.5(p=7.8x10-19) and OR 1.13 (p=6.1x10-25) in Sikhs and SouthAsians, respectively [6]. However, despite extensive replication, no study has unequivocallydemonstrated the underlying molecular mechanism of this association. Little is knownabout the clinical role of TCF7L2 in T2D beyond progression from impairedglucose tolerance to diabetes [7].
Various in vitro and in vivo studies have shown that severalcomponents of the Wnt pathway are involved in β-cell proliferation [8], insulin secretion and cholesterol metabolism [9], and production of glucogon-like peptide-1 (GLP-1) [10]. Wnts are secreted glycoproteins with a well-established role in theearly stages of development through adulthood [11]. Wnts bind to frizzled and LRP receptors, which, in turn, inactivate thedegradation complex consisting of AXIN, DVL, and GSK3B (Figure 1). This prevents the phosphorylation of β-catenin by GSK3B, andleads to its binding to the nuclear transcription factors, TCF7, LEF1, TCF7L1 andTCF7L2, leading to the activation of more than 60 different genes involved in growthregulation and differentiation, as well as GLP-1 expression [12]. Since Wnt signaling has a role in regulating and stabilizingβ-catenin and its binding with TCF7L2, we hypothesized that any alternation inthe canonical Wnt pathway would have profound consequences in insulin secretion andthe generation of new β-cells, particularly given that Wnt signaling isrequired for normal development of the pancreas and islets during embryonic growth [13].
The present investigation is a follow-up study to explore the role of a novel,four-nucleotide (CCCA) insertion polymorphism we discovered in the most conservedregion of WNT16a in US American Sikhs. The objectives of this investigationare: 1) to study the potential role of this WNT16a insertion in T2D in ourdiabetic sample of Punjabi Sikhs, 2) to quantify and compare gene expression ofWNT16a and TCF7L2 between carriers and non-carriers of theCCCA insertion within the WNT16a gene using mRNA samples from 27 frozenhuman pancreatic tissues, 3) to investigate the functional impact of this insertionon protein levels and message translation using a luciferase reporter vectorcontaining the wild-type and mutant WNT16a 5′untranslated regions (UTR)transfected into cultured cells, and 4) to perform immunohistochemistry to examinethe expression of WNT16a in human pancreas among insertion carriers vs.non-carriers.
Methods
Study participants
The DNA samples of 2,034 (52% male) individuals from our ongoing Sikh DiabetesStudy (SDS) were used [14]. Of these, ~48% were ascribed as having T2D based on establishedguidelines of the American Diabetes Association, as described [15]. A medical record indicating either (1) a fasting blood glucose (FBG)≥126 mg/dL (≥7.0 mmol/L) after aminimum 12 h fast or (2) a 2 h post-glucose level (2 h oralglucose tolerance test [OGTT]) ≥ 200 mg/dL(≥11.1 mmol/L) on more than one occasion, combined withsymptoms of diabetes, confirmed the diagnosis. Impaired fasting glucose (IFG) isdefined as a fasting blood glucose level ≥100 mg/dL(5.6 mmol/L) but ≤126 mg/dL (7.0 mmol/L), asdescribed previously [16]. Common characteristics observed in diabetics include excessivethirst, hunger, polyuria, blurry vision, common skin and urinary tractinfections, nocturia, loss of bladder control, and fatigue. Impaired glucosetolerance (IGT) is defined as a 2 h OGTT >140 mg/dL(7.8 mmol/L) but <200 mg/dL (11.1 mmol/L). Subjects with IFGor IGT were considered pre-diabetics and were excluded from the analysis. The2 h OGTTs were performed following the criteria of the World HealthOrganizations (WHO) (75 g oral load of glucose). Body mass index (BMI) wascalculated as (weight (kg)/height (meter) [2]. Homeostasis Model Assessment (HOMA) for insulin resistance (HOMA-IR)was calculated as fasting glucose X fasting insulin/22.5, as described [17].
The normoglycemic subjects were recruited from the same Punjabi Sikh communityand geographic location as the T2D patients [14]. The majority of the subjects were recruited from the state of Punjabin North India and Punjabi Sikhs living in the US. Individuals of South, East,and Central Indian origin were excluded, as were individuals with type-1diabetes, a family member with type 1 diabetes, rare forms of T2D calledmaturity-onset diabetes of young (MODYs), or secondary diabetes (e.g.,hemochromatosis, pancreatitis). Demographic and clinical characteristics of theSDS subjects are summarized in Table 1. All blood samples were obtained at thebaseline visit and all participants provided a written informed consent forthese investigations. All SDS protocols and consent documents were reviewed andapproved by the University of Oklahoma Institutional Review Board and the HumanSubject Protection Committees at the participating hospitals and institutes inIndia.
Metabolic estimations
Insulin was measured by radio-immuno assay (Diagnostic Products, Cypress, USA).Serum lipids (total cholesterol, low density lipoprotein cholesterol [LDL-C],high-density lipoprotein [HDL-C], very low-density lipoprotein cholesterol[VLDL-C], and triglycerides [TG]) were measured by using standard enzymaticmethods (Roche, Basel, Switzerland), as described [16, 18]. C-peptide, TNFα, and MCP-1 measures were simultaneouslyquantified using Millipore’s Magnetic MILLIPLEX Human Metabolic panel (St.Charles, MO) and analyzed on a Bio-plex 200 multiplex system (Bio-Rad Hercules,CA), as described previously [19].
Whole-genome exome sequencing
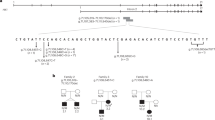
We performed genome-wide exome sequencing on two Punjabi Sikh subjects: a64-year-old healthy normoglycemic male, and a 67-year-old diabetic female, usingan Illumina GAIIx and “SureSelect Human All Exon Kit” by AgilentTechnologies and “Paired-End Sequencing Library Prep by Illumina”(Version 1.0.1). The sequences containing 75x reads were filtered against publicdatabases of genetic variants. The present investigation is focused on exploringthe role of a frame-shift insertion (CCCA) discovered in a conserved region ofhuman WNT16a gene (Additional file 1: FigureS1).
Genotyping
Genotyping of the insertion polymorphisms was performed by polymerase chainreaction (PCR) and a gel-based assay. Forward primer Wnt16a-F (5')[TACCACTCTCCTCCCTCC] and reverse primer Wnt16a-R (3') [CCCTGATCAAATCCCCAAAT]were used to amplify the region containing the identified insertion; PCRamplification generated a 458 bp product in the sample containing noinsertion. PCR conditions included an initial denaturation for 5 min. at95°C, followed by 36 cycles (30 sec. 95°C, 45 sec.53.7°C, 30 sec. 72°C), and a 10 min. extension at 72°C.Positive and negative controls were included for every PCR. 15μL of the PCRproduct was then separated on a 2.5% nusieve/agarose gel (3:1) for2.5 hours at 140 volts to determine the genotype of participants asinsertion (462 bp), non-insertion carriers 458 bp, and heterozygotescontaining insertion/normal sequence of 462/458 bp (Additional file 1: Figure S2). To confirm the presence of theWNT16a insertion scored on the gel-based assay, approximately 30samples were sequenced using an ABI 3730 capillary sequencer (Applied BiosystemsInc. Foster City, CA) and were analyzed using Mutation Surveyor DNA variantanalysis software (v4.0.6.)(SoftGenetics, State College, PA). Genotypingof rs7903146, located in intron 3 of the TCF7L2 gene, was performedwith a TaqMan genotyping assay (Applied Biosystems, Foster City, CA), using a7900 genetic analyzer, as described previously [4].
Quantitative gene expression studies on WNT16a
Gene expression studies for Wnt16a were performed using 27 human pancreatictissue specimens (13 diabetic and 14 non-diabetics) collected from theDepartment of Surgery at the University of Oklahoma Health Sciences Center.Total RNA was extracted from frozen tissues (stored in liquid nitrogen) usingAmbion’s mirVana RNA kits (Grand Island, NY), followed by RT-PCR usingBio-Rad’s iScript RT-PCR kit (Hercules, CA), according to themanufacturers’ instructions. Real Time PCR was then performed using an ABI7900HT genetic analyzer in conjunction with Qiagen’s QuantiTect primerassay (Chatworth, CA) and Bio-Rad’s iTaq SYBR Green Supermix with ROX(Hercules, CA). Results were then analyzed on ABI’s RQ Manager (v.1.2.1)software. Beta-actin was used as a normalizing control.
Transient DNA transfection and dual-luciferase assay
The 5′ UTRs of the wild-type and mutant Wnt16a message were incorporatedinto oligonucleotide primers as depicted in Additional file 1: Figure S3. Note that each of the 5′ primers incorporateda Sac I site for insertion into pCI-GFP, followed by the sequence of the Wnt16a5′ UTR, then a region homologous to firefly luciferase. The pCI-GFP vectorwas developed by inserting eGFP into the parent vector, pCI-Neo (Promega,Madison, WI), and allowed us to monitor transfection efficiency. The 3′primer was homologous to a site in the pGL3 vector past a unique Xba I site inthe vector. After PCR amplification using pGL3 as a template, the amplimers weredigested with Sac I and Xba I, and then ligated into pCI-GFP. For transfectioninto cultured cells, each construct (0.125 μg per culture well) wasadded to 1 μl Plus reagent and 15 μl Opti-MEM (LifeTechnologies, Carlsbad, CA), along with 0.125 μg per well of an emptypGL3-Basic vector (which served as carrier DNA) and 0.01 μg per wellpGL4.74 (a Renilla luciferase construct used for normalization) for a total of0.26 μg DNA. This was added to 0.5 μl Lipofectamine reagentin an additional 15 μl of Opti-MEM and used to transfect HEK-293 cells(74,000 cells per well) in a 48-well plate. After 48 hours in medium plus10% calf serum, cells were washed in PBS, and lysed for luciferase activity.Lysates were diluted until the luciferase values fell within a linear responserange. Both firefly and Renilla luciferase values were measured using adual luciferase detection kit (Promega, Madison, WI).
Immunohistochemistry
Formalin-fixed paraffin-embedded pancreatic tissues were cut at a thickness of4 μm, mounted on SuperfrostPlus® slides (Statlab MedicalProducts, Lewisville, TX), and subsequently deparaffinized, rehydrated, andwashed in Tris Buffered Immunohistochemistry wash buffer + Tween 20 (TBST,catalog# 935B, Cell Marque, Rocklin, CA). Antigen retrieval was accomplished byplacing slides in 10 mM citrate buffer, pH 6.0 (cat. #S2389,TargetRetrieval Solution, DAKO, Carpentaria, CA), in a steamer for 20 minutes,followed by 20 minutes cooling in deionized water at room temperature.According to the manufacturer’s directions, sections were treated with abackground blocker (cat. #927B, Cell Marque, Rocklin, CA) and a peroxidaseblocking reagent (cat. #925B, Cell Marque, Rocklin, CA) to inhibit endogenousperoxidase activity, followed by three, five-minute washes each in deionizedwater. Rabbit anti-Wnt antibody was prepared in antibody diluent (cat. #936B,Cell Marque, Rocklin, CA) and added to slides at 2 μg/ml (1:500dilution, cat. #LS-A9630, MBL International Corporation, Woburn, MA). Antigenretrieval was accomplished according to the manufacturer’s recommendationfor the Wnt16 antibody (LSBio, Woburn, MA). Following incubation for 1 hrat room temperature, the sections were processed for immunohistochemistry usingthe HiDef detection HRP Mouse/Rabbit polymer system (cat. #954D, Cell Marque,Rocklin, CA). Sections were washed three times for five minutes each in trisbuffered immunohistochemistry wash buffer + Tween 20 (TBST), incubated with theamplifier, washed three times for five minutes each in TBST, and incubate withlabeled polymer. Following a final wash in TBST, slides were incubated with3′3′diaminobenzidine tetrahydrochloride (DAB) (cat. #957D, DABsubstrate Kit, Cell Marque, Rocklin, CA). Counterstaining was performed withImmuno* Master Hematoxylin (American Master*Tech Scientific,Inc., Lodi, CA). Controls were incubated with rabbit IgG isotype at2 μg/ml (rabbit [DA1E] mAB IgGXP® isotype control, cat. #3900,Cell Signaling Technologies Danvers, MA). A total of seven tissues (1 T2D and 6controls) with Wnt16a genotypes were used for immunohistochemistry.Slides were scored based on intensity (0- no, 1- weak, 2- moderate and 3-strong), and the area of stain (0 for 0%, 1-<10%, 2-between 10-15%, and 3-between 51-81%). The consolidated scores (ranging from 0–7) were derivedfrom the sum of scores of intensity and area, negative being in the range of0–2, weakly positive-3, moderately positive ranging from 4–5, andhighly positive ranging from 6–7.
Statistical analysis
Association analysis
Data quality for SNP genotyping was checked by establishing reproducibilityof control samples. Departure from Hardy-Weinberg equilibrium in controlswas checked using Pearson’s Chi-square, as reported previously [5]. Descriptive statistical analyses were performed with SPSSStatistics Software (v 15.0). The chi-square test for categorical variablesand t-test for continuous variables were used to test differences whereappropriate. While multivariate logistic-regression was used to assess theassociation of the insertion with T2D and obesity, multivariatelinear-regression was used for each quantitative trait after adjustment forrelevant covariates (age, sex, diabetes status, BMI, and medication),assuming an additive model. Skewed variables were detected byShapiro-Wilk’s test for continuous traits. Subsequently, TG, totalcholesterol, LDL-C, VLDL-C, FBG, C-peptide, MCP-1, and HOMA-IR werenormalized by log-transformation before statistical comparisons, and allp-values were derived from analyses of transformed data. The summarystatistics (β, S.E., and p-values) were used to assess SNP-phenotypeassociation. Gene expression analyses were performed using AppliedBiosystems’ RQ Manager (v.1.2), which uses the comparativeCT method for relative quantification. We determined theΔCT value by (Target Average CT-EndogenousControl Average CT), then calculated theΔΔCT to determine the fold-difference in geneexpression by ΔCT Target - ΔCT Calibrator.For the amount of target determination, the data were normalized to theendogenous control and relative to the calibrator by using2-ΔΔCT as described [20] . For reporter assays, the results are presented as the mean± average deviation from the mean for the number of observation, asindicated. Statistical significance of differences between groups wasestimated using a two-tailed t test.
Results
Whole-exome sequencing
As summarized in Additional file 2: Table S1, a totalof 20,306 mutations were found in the control and 21,258 in the diabeticsubjects. Among these, 4,673 and 4,842 novel SNPs were uniquely present incontrol and T2D cases, respectively. To identify the functional significance ofthe variants identified, we performed initial comparative genomic screening onthe mutations found in some selected loci using UCSC’s Vista GenomeBrowser. From these results, several candidate genes involved in insulinsecretion, β-cell proliferation, or related pathways were identified (datanot shown). Interestingly, novel substitution in WNT16a, which showed a4-base-pair frame-shift insertion near two known SNPs, was in an evolutionarilyconserved region (as shown in Additional file 1:Figure S3) and was predicted to be disruptive.
Association studies
A genetic screening of 2,034 SDS individuals (977 T2D cases and 1,057 controlsT2D cases and 1,057 controls) showed that 33% of T2D cases and 32% non-diabeticcontrols were carriers of a CCCA insertion; the number of carriers of thisinsertion did not differ significantly among cases versus controls (p=0.08).Multiple regression analysis, performed in diabetics and non-diabetic controlsseparately, did not reveal any association of the Wnt16a insertion withobesity (BMI, waist-to-hip ratio [WHR]) (Table 1).However, the insertion carriers showed moderately higher mean (±SD) levelsof total cholesterol compared to non-carriers (173.8±52.9 (mg/dL) vs.179.2±49.9 (mg/dL), p=0.038). Serum mean levels of inflammatory cytokinesTNFα were also significantly higher among insertion carriers compared tonon-carriers (p=0.008) (Figure 2). A similar butnon-significant trend was seen with increased mean levels of MCP-1 amonginsertion carriers compared to non-carriers (p=0.440) (Figure 2). There was a significant difference in the frequency of‘T’ (the at risk allele for T2D) in rs7903146 of TCF7L2among cases and controls (38% cases vs. 28% controls). The age- and sex-adjustedOR showing ‘T’ allele-associated T2D risk was 1.51 (95%CI[1.37-1.66], p=1.53x10-17). However, no association ofTCF7L2 polymorphism was seen with inflammatory cytokines (TNFαor MCP-1) (data not shown).
Distributions of serum levels (mean ±SD) of inflammatorycytokines (TNFα and MCP-I) among Wnt16a insertion carriersnon-carriers in SDS subjects. Serum levels of TNFα weresignificantly higher (p=0.008) in insertion carriers, while a similarbut non-significant trend was seen with MCP-1. The statistical analysiswas performed in combined samples (T2D and controls) after adjusting forthe confounding effects of age, BMI, gender, and T2D status.
Bioinformatics, gene expression studies, and western blotting
The Wnt16a message, which is uniquely expressed in pancreas, includes an upstreamopen reading frame (uORF) that initiates 14 bp 5′ of the codingsequence AUG (Figure 3). Translation of this 5′UTR would terminate 140 bp later, presumably resulting in nonsense-mediateddecay (NMD) of the message, since two down-stream exon junction complexes wouldnot be disrupted during the pioneer round of translation [21]. The 4-base-pair insertion (CCCA) of the mutated Wnt16a message, onthe other hand, would result in the transition of the uORF to an in-framealternative translation initiation site. In this case, translation initiationfrom either the first or second AUG during the pioneer round of translationwould not trigger NMD. RT-PCR and qualitative gene expression studies wereperformed by quantifying mRNA expression of WNT16a and TCF7L2genes among carriers and non-carriers of Wnt16a. Of 27 participant donors ofhuman pancreatic tissue, nine were carriers of the insertion in WNT16a.As shown in Figure 4, our data revealed a ~3.2-foldincrease in the expression of WNT16a among insertion carriers compared tonon-carriers. The expression of WNT16a was consistently higher among insertioncarriers irrespective of disease status. Gene expression analysis ofTCF7L2 in the same pancreatic tissues revealed a significantelevation (p=0.003) of the amount of TCF7L2 mRNA among insertion carrierscompared to non-carriers (Figure 5A). In thestratified data by disease within CCCA insertion carriers, the expression ofTCF7L2 mRNA was higher in diabetic pancreatic tissues compared to non-diabeticpancreas, however, this increase was not statistically significant (p=0.155).(Figure 5B). Further stratification ofquantitative mRNA expression among the at-risk ‘T’ allelecarriers of TCF7L2 SNP (rs7903146) revealed that the CT+TT genotypesshowed an 8.7-fold increase in the expression of TCF7L2 compared to CC genotypesin Wnt16 a insertion carriers, while the non-insertion carriers showed the sameallelic trend at a significantly reduced magnitude (Figure 6).
The human Wnt16 gene includes two alternativetranscription start sites, resulting in two alternative first exonsand three common exons. The Wnt16a message, which is onlyexpressed in pancreas, includes an upstream open reading frame (uORF)that initiates 14 bp 5′ of the coding sequence AUG. As shown,in the Wnt16a wild-type allele, translation of the 5′ UTRwould terminate 140 base pairs later, presumably resulting innonsense-mediated decay, since two down-stream exon junction complexeswould not be disrupted during the pioneer round of translation. The 4base-pair CCCA insertion of the mutated Wnt16a message, on the otherhand, results in the transition of the uORF to an in-frame alternativetranslation initiation site. Translation initiation from either AUGduring the pioneer round of translation would not triggernonsense-mediated decay of the Wnt16a massage.
Gene expression studies for Wnt16a were carried out using 27 pancreastissue samples by quantifying mRNA expression of WNT16a by real-timePCR. Of 27 participant donors of human pancreatic tissue, ninewere carriers of CCCA insertion in WNT16a. Our data revealed a3.2 fold increase in the expression of WNT16a among insertion carrierscompared to non-carriers.
Gene expression study of TCF7L2 in the same 27 pancreatic tissues usedto determine the expression of Wnt16a by real-time PCR analysis.Figure 5A shows a significant elevationof TCF7L2 mRNA levels among CCCA insertion carriers and a very lowexpression of TCF7L2 mRNA was observed in non-carriers (p= 0.003).Figure 5B shows that within CCCAinsertion carriers, the expression of TCF7L2 mRNA in pancreas waselevated among diabetics compared to non-diabetic controls.
Stratification of TCF7L2 mRNA quantitation by TCF7L2 genotypes of rs7903146 among Wnt16a insertion carriersand non-carriers. The at-risk ‘T’ allele carriers ofTCF7L2 with CT+TT genotypes showed a 8.7-fold increase inthe expression of TCF7L2 compared to CC genotypes. Note that thisincrease was only observed in Wnt16a (CCCA) insertion carriersand not in the non-carriers.
Luciferase reporter assay
In order to further evaluate the influence of the CCCA insertion on translationof the Wnt16a message, we assembled reporter constructs driven by thecytomegalovirus (CMV) promoter that included the wild-type and the mutantsequence of the Wnt16a 5′ UTRs. The long uORF was mimicked in ourluciferase construct by the presence of a translation stop site in-frame withthe upstream AUG. If the first AUG in the message was used to initiatetranslation, then no luciferase protein should have been produced. Indeed, whenwe inserted the additional four nucleotides to replicate what occurs in themutant situation, we noted a significantly increased level (~70%) of luciferaseexpression (p=0.0001) (Figure 7). This suggests thatthe upstream AUG can act as an efficient translation initiation site. In thewild-type gene, this would reduce expression of the full-length protein bypreventing initiation at the second AUG. In the presence of the CCCA insertion,both AUGs are in the same reading frame, so full-length protein would beproduced regardless of which AUG was used to initiate translation.
Reporter constructs assembled with the mutated form of the Wnt16a5′ UTR are more efficiently translated than the wild-typeform. The wild-type and mutant Wnt16a 5′ UTR sequenceswere inserted adjacent to a luciferase cDNA, and the resulting plasmidswere used to transfect HEK-293 cells. 48 hours after transfection,cells were lysed, and firefly luciferase (FFL) and Renillaluciferase (RL) levels were measured. Our reporter constructs using thewild-type and the mutant (insertion) sequence of Wnt16a showedsignificantly increased (70%, p<0.0001) levels of luciferase proteinin the constructs carrying the mutant sequence.
Immunohistochemistry
Immunoperoxidase staining of paraffin-embedded pancreatic tissues ofnormoglycemic controls and diabetic cases were scored for the intensity ofantibody as described in methods. As shown in Figure 8, the tissues with insertion carriers revealed a higher expressionof Wnt16a showing high intensity staining among insertion carriers versesnegative staining in non-carriers. The scoring intensity was indifferent amongdiabetics and non-diabetics.
Immunoperoxidase staining of 5 micron thick histological sections ofparaffin-embedded pancreatic tissues. The sections wereprocessed for immunohistochemistry using the HiDef detection HRPMouse/Rabbit polymer system as described in detail under methods. TheCCCA insertion carriers revealed a higher expression of Wnt16, showingstaining score from +1 to +3 among insertion carriers versus negativestaining in non-carriers, which showed staining scores from 0 to +1. Thescoring intensity was not different among diabetic andnon-diabetics.
Discussion
The key effector pathway of Wnt signaling (β-cat/TCF7L2) has beenrecently implicated in metabolic homoeostasis, diabetes, obesity, osteoporosis,cardiovascular disease, and cancer [9, 22–24]. The discovery of TCF7L2 as a T2D susceptibility gene indifferent ethnic populations through genome-wide studies has triggered numerousinvestigations to explore the clinical utility of identifying TCF7L2genetic variations, and whether the identified SNPs can be used as markers fortailoring customized therapeutics. However, the underlying molecular mechanism bywhich TCF7L2 variants influence T2D remains unclear. While a number ofrecent studies have suggested the essential involvement ofβ-cat/TCF7L2 in the Wnt signaling pathway for pancreatic developmentand function [25, 26], the role of β-cat in pancreatic β celldevelopment remains unclear and controversial [13, 27]. Mice lacking β-cat developed pancreatitis prenatally;however, they later recovered from pancreatitis and regenerated normal pancreas andduodenal villi from wild-type cells that escaped earlier β-catdeletion. These observations suggested that mouse embryos were capable of overcomingsubstantial β-cat reduction through complicated compensatorymechanisms [13]. Other studies have shown that the over-expression of β-catat different development stages generated different effects [27]. Similarly, some studies suggest an essential and beneficial role ofTCF7L2 in pancreatic β cell development [28, 29], while other studies revealed a destructive role of TCF7L2 byover-expression of TCF7L2 mRNA due to alternatively spliced variants, whichincreased the risk of developing T2D [30]. Further, the increased expression of TCF7L2 in pancreatic β-cellswas positively correlated with insulin gene expression but was negatively correlatedwith glucose-stimulated insulin release [30]. Therefore, it is still unclear how β-cat/TCF in Wntsignaling is mechanistically involved in pancreatic development and increased T2Dsusceptibility.
In this investigation, the discovery of a frame-shift insertion in the most conservedregion of WNT16a (Additional file 1: Figure S4),and the restricted and exclusive expression of Wnt16a isoform in the human pancreas [31], prompted us to explore the role of this Wnt16a insertion in T2Dusing genetic epidemiologic, molecular, and physiologic studies. TCF7L2polymorphisms have demonstrated the biggest effect on the risk for developing T2D inrecent GWAS and replication studies in multiple ethnic populations, including ourown studies in Asian Indians [4–6, 32, 33]. The Wnt16a isoform is exclusively expressed in the pancreas ofhumans, while its close relative, Wnt16b, is ubiquitously expressed in many otherorgans [31]. The prevalence of the CCCA insertion polymorphism did not differsignificantly among diabetic cases (33%) versus controls (32%) in our cohort.Although our epidemiological data did not clarify the role of CCCA insertion in T2D,obesity, or lipid metabolism (Table 1), our multiple linear regression resultsshowed significant elevation in serum TNFα levels among insertion carriersversus non-carriers (p= 0.008), as well as a non-significant trend in the samedirection for another inflammatory marker, MCP-1 (p=0.44). These findings are inagreement with earlier studies reporting the influence of Wnt signaling ininflammation [34], and suggest that the presence of the CCCA insertion appears to promotecirculatory levels of pro-inflammatory cytokines in our samples.
Our in silico analysis (Figure 3) clearlysuggested that the frame-shift insertion of the mutated WNT16a results in thetransition of the uORF to an in-frame alternative translation initiation site.During the pioneer round of translation, initiation at this up-stream AUG would notresult in NMD. In non-carriers, initiation at this up-stream AUG would prevent theproduction of mature protein, and would likely result in NMD, thereby reducing theexpression of this gene. This was further verified in our quantitative real-time PCRresults that consistently showed the wild-type (non-insertion carriers) messagelevels being ~3.2-fold lower than those observed in samples from the insertioncarriers (Figure 4). Additional evidence of the influenceof the CCCA insertion on translation of the message was obtained using reporterconstructs that incorporated the wild-type and the mutant (insertion) sequence ofthe WNT16a 5′ UTR. Using this approach, we noted a marked increase in thelevels of luciferase expression in the constructs carrying insertion (p=0.0001)(Figure 7). This was additionally confirmed inhistological sections of the embedded human pancreatic islets stained with Wnt16antibody. It was interesting to observe that the tissues with insertion carriersshowed higher expression of Wnt16a with staining score ranging from +1 to +3 versesnegative staining in non-carriers (Figure 8).
Our comparison of the expression of TCF7L2 mRNA in the same pancreatic tissues usedfor Wnt16a analysis showed a significantly increased (p=0.003) expression of TCF7L2among the WNT16a insertion carriers compared to the wild-type(non-carriers) (Figure 5A). This significantly enhancedexpression of Wnt16a and TCF7L2 among insertion carriers in human pancreas would bepredicted to affect the expression of several β-cat /TCF7L2 or Wntdownstream target genes [22]. It was interesting to observe that, despite the fact that the frequencyof the at-risk ‘T’ allele in rs7903146 of TCF7L2 did not differamong WNT16a insertion and non-carriers (0.34 insertion carriers vs. 0.33non-carriers), TCF7L2 mRNA levels were significantly elevated (~23 folds) amongWNT16a insertion carriers vs. non-carriers (Figure 5A). Additionally, the at-risk ‘T’ allele carriers ofTCF7L2 (rs7903146) also showed significantly increased expression ofTCF7L2 mRNA in pancreas compared to CT and CC carriers (Figure 6). This is consistent with enhanced Wnt signaling, something we wouldpredict given the impact of the Wnt16a insertion mutation identifiedhere.
TCF7L2 has been shown to be abundantly expressed in GLP-1-producing intestinalepithelial cells [35]. It has also been shown to be expressed in pancreas and to mediatepancreatic β cell proliferation and survival [28, 36]. However, in other studies, TCF7L2 was shown to be present at low levelsor not expressed at all in pancreas [29, 35, 37]. We have identified a significant elevation of TCF7L2 mRNA in pancreas,especially among the CCCA insertion carriers, which appears to increase diabetesrisk by increasing the expression of TCF7L2 among ‘T’ risk allelecarriers of rs7903146 of TCF7L2. These results suggest a synergistic effectof Wnt16a insertion and the at-risk ‘T ’allele ofTCF7L2 in compounding the risk of T2D, likely through elevatedβ-cat/TCF7L2 activity and the expression of downstream Wnt targets. Higherexpression of TCF7L2 among ‘T’ allele carriers was evident in pancreatictissues of diabetic patients compared to non-diabetic controls. These results are inagreement with earlier findings by Lysenko et al. [30], where carriers of ‘T’ allele in rs7903146 of TCF7L2exhibited five-fold increases in TCF7L2 mRNA levels in pancreatic islets of diabeticpatients, and showed an associated impairment of insulin secretion. Previousfindings by others have shown that, while elevated mRNA expression of TCF7L2 waslinked with ‘T’ risk allele of rs7903146, even though no apparentincrease in TCF7L2 protein amount was observed [38, 39]. In spite of this, the same groups demonstrated that the higher mRNAexpression of TCF7L2 variants resulted in the down-regulation of GLP-1-inducedinsulin secretion, and increased the risk of T2D through Wnt signaling [38, 40]. Since GLP-1 receptors are primarily located in pancreas and Wnt16a isexclusively expressed in pancreas, it is quite conceivable common insertionpolymorphism in WNT16a may affect GLP-1 receptor activity by modulatingTCF7L2 expression, thus influence GLP-1-induced insulin secretion. Since Wntsignaling is known to stabilize the binding of β-catenin with TCF7L2, which iscritical for expression of many other genes involved in β-cell development, anyalteration in the canonical Wnt pathway should have profound consequences in insulinsecretion and the generation of new β-cells, as this pathway is required to betightly regulated. It will be also of interest to determine if WNT16a can modulateGLP-1 receptor expression independent of TCF7L2.
Conclusions
To our knowledge, ours is the first study reporting the role of WNT16a inβ-cat/TCF7L2 signaling and the risk of developing T2D, whichappears to be mediated through the increased expression of TCF7L2 in pancreas, apathway critical for the regulation of several dozen downstream genes involved inglucose metabolism, apoptosis, skeletal muscle function, and atherosclerosis.Therefore, a detailed examination of Wnt16a and its potential role in geneticpredisposition to T2D through Wnt signaling, and cross-talk between other signalingpathways, may help identify therapeutic targets for the treatment of T2D.
Author contributions
Conceived and designed the experiments: DKS; Provided pancreatic tissues andimmunohistochemistry: DB, ML, SL; Western blotting and luciferase studies: EWH, ECB;Genotyping, gene expression and analysis: LFB; Contributedreagents/materials/analysis tools: DKS and EWH; Wrote the paper: DKS and EWH;Guarantors: DKS, EWH. All authors read and approved the final manuscript.
References
Grant SF: Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2diabetes. Nat Genet. 2006, 38: 320-3. 10.1038/ng1732.
Ip W, Chiang YT, Jin T: The involvement of the wnt signaling pathway and TCF7L2 in diabetes mellitus:The current understanding, dispute, and perspective. Cell Biosci. 2012, 2: 28-10.1186/2045-3701-2-28.
Cauchi S: TCF7L2 is reproducibly associated with type 2 diabetes in various ethnicgroups: a global meta-analysis. J Mol Med (Berl). 2007, 85: 777-82. 10.1007/s00109-007-0203-4.
Sanghera DK: TCF7L2 polymorphisms are associated with type 2 diabetes in Khatri Sikhs fromNorth India: genetic variation affects lipid levels. Ann Hum Genet. 2008, 72: 499-509. 10.1111/j.1469-1809.2008.00443.x.
Sanghera DK: Impact of nine common type 2 diabetes risk polymorphisms in Asian IndianSikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer asignificant risk. BMC Med Genet. 2008, 9: 59-
Saxena RS SD, Been LF, Gravito ML, Braun T, Bjonnes A, Young R, Ho W, Rasheed A, Frossard P, Xueling S, Hasnali N, Venkatesan R, Chidambaram M, Liju S, Rees S, Peng-Keat Ng D, Wong TY, Yamauchi T, Hara K, Tanaka Y, Hirose H, McCarthy M, Morris A, Basit A, Barnett A, Katulanda P, Matthews D, Mohan V, Wander GS, Singh JR, Mehra N, Ralhan S, Kamboh MI: Genome-wide association study identifies novel loci contributing to type 2diabetes in individuals of Punjabi origin from Southeast Asia. Diabetes. 2013, 10.2337/db12-1077..
Florez JC: TCF7L2 polymorphisms and progression to diabetes in the Diabetes PreventionProgram. N Engl J Med. 2006, 355: 241-50. 10.1056/NEJMoa062418.
Polakis P: Wnt signaling and cancer. Genes Dev. 2000, 14: 1837-51.
Fujino T: Low-density lipoprotein receptor-related protein 5 (LRP5) is essential fornormal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA. 2003, 100: 229-34. 10.1073/pnas.0133792100.
Doble BW, Woodgett JR: GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003, 116: 1175-86. 10.1242/jcs.00384.
Moon RT, Kohn AD, De Ferrari GV, Kaykas A: WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004, 5: 691-701. 10.1038/nrg1427.
Gordon MD, Nusse R: Wnt signaling: multiple pathways, multiple receptors, and multipletranscription factors. J Biol Chem. 2006, 281: 22429-33. 10.1074/jbc.R600015200.
Papadopoulou S, Edlund H: Attenuated Wnt signaling perturbs pancreatic growth but not pancreaticfunction. Diabetes. 2005, 54: 2844-51. 10.2337/diabetes.54.10.2844.
Sanghera DK: The Khatri Sikh Diabetes Study (SDS): study design, methodology, samplecollection, and initial results. Hum Biol. 2006, 78: 43-63. 10.1353/hub.2006.0027.
American Diabetes Association: Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004, 27 (Suppl 1): S5-S10.
Sanghera DK: Testing the association of novel meta-analysis-derived diabetes risk geneswith type II diabetes and related metabolic traits in Asian Indian Sikhs. J Hum Genet. 2009, 54: 162-8. 10.1038/jhg.2009.7.
Matthews DR: Homeostasis model assessment: insulin resistance and beta-cell function fromfasting plasma glucose and insulin concentrations in man. Diabetologia. 1985, 28: 412-9. 10.1007/BF00280883.
Sanghera DK: Genome-wide linkage scan to identify loci associated with type 2 diabetes andblood lipid phenotypes in the Sikh Diabetes Study. PLoS One. 2011, 6: e21188-10.1371/journal.pone.0021188.
Braun TR,BL, Blackett PR, Sanghera DK: Vitamin D deficiency and cardio-metabolic risk in a north indian communitywith highly prevalent type 2 diabetes. J Diabetes Metab. 2012, 3: 213-
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCRand the 2(−Delta Delta C(T)) Method. Methods. 2001, 25: 402-8. 10.1006/meth.2001.1262.
Maquat LE, Hwang J, Sato H, Tang Y: CBP80-promoted mRNP rearrangements during the pioneer round of translation,nonsense-mediated mRNA decay, and thereafter. Cold Spring Harb Symp Quant Biol. 2010, 75: 127-34. 10.1101/sqb.2010.75.028.
Jin T, Liu L: The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol. 2008, 22: 2383-92. 10.1210/me.2008-0135.
Rachner TD, Khosla S, Hofbauer LC: Osteoporosis: now and the future. Lancet. 377: 1276-87.
Manolagas SC, Almeida M: Gone with the Wnts: beta-catenin, T-cell factor, forkhead box O, andoxidative stress in age-dependent diseases of bone, lipid, and glucosemetabolism. Mol Endocrinol. 2007, 21: 2605-14. 10.1210/me.2007-0259.
Lim HW: Identification of differentially expressed mRNA during pancreas regenerationof rat by mRNA differential display. Biochem Biophys Res Commun. 2002, 299: 806-12. 10.1016/S0006-291X(02)02741-9.
Rulifson IC: Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci USA. 2007, 104: 6247-52. 10.1073/pnas.0701509104.
Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M: Stabilization of beta-catenin impacts pancreas growth. Development. 2006, 133: 2023-32. 10.1242/dev.02366.
Shu L: Transcription factor 7-like 2 regulates beta-cell survival and function inhuman pancreatic islets. Diabetes. 2008, 57: 645-53. 10.2337/db07-0847.
Korinek V: Depletion of epithelial stem-cell compartments in the small intestine of micelacking Tcf-4. Nat Genet. 1998, 19: 379-83. 10.1038/1270.
Lyssenko V: Mechanisms by which common variants in the TCF7L2 gene increase risk of type2 diabetes. J Clin Invest. 2007, 117: 2155-63. 10.1172/JCI30706.
Fear MW, Kelsell DP, Spurr NK, Barnes MR: Wnt-16a, a novel Wnt-16 isoform, which shows differential expression in adulthuman tissues. Biochem Biophys Res Commun. 2000, 278: 814-20. 10.1006/bbrc.2000.3852.
Kooner JS: Genome-wide association study in individuals of South Asian ancestryidentifies six new type 2 diabetes susceptibility loci. Nat Genet. 43: 984-9.
Mao H, Li Q, Gao S: Meta-analysis of the relationship between common type 2 diabetes risk genevariants with gestational diabetes mellitus. PLoS One. 2012, 7: e45882-10.1371/journal.pone.0045882.
Gustafson B, Smith U: Cytokines promote Wnt signaling and inflammation and impair the normaldifferentiation and lipid accumulation in 3 T3-L1 preadipocytes. J Biol Chem. 2006, 281: 9507-16.
Yi F, Brubaker PL, Jin T: TCF-4 mediates cell type-specific regulation of proglucagon gene expressionby beta-catenin and glycogen synthase kinase-3beta. J Biol Chem. 2005, 280: 1457-64.
Liu Z, Habener JF: Glucagon-like peptide-1 activation of TCF7L2-dependent Wnt signaling enhancespancreatic beta cell proliferation. J Biol Chem. 2008, 283: 8723-35. 10.1074/jbc.M706105200.
Barker N: Identification of stem cells in small intestine and colon by marker geneLgr5. Nature. 2007, 449: 1003-7. 10.1038/nature06196.
Schafer SA: Impaired glucagon-like peptide-1-induced insulin secretion in carriers oftranscription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007, 50: 2443-50. 10.1007/s00125-007-0753-6.
Shu L: Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate withdownregulation of GIP- and GLP-1 receptors and impaired beta-cellfunction. Hum Mol Genet. 2009, 18: 2388-99. 10.1093/hmg/ddp178.
Schafer SA, Machicao F, Fritsche A, Haring HU, Kantartzis K: New type 2 diabetes risk genes provide new insights in insulin secretionmechanisms. Diabetes Res Clin Pract. 2011, 93 (Suppl 1): S9-24.
Acknowledgements
This work was partly supported by R01DK082766 funded by the National Institute ofDiabetes and Digestive and Kidney Diseases and NOT-HG-11-009 funded by NationalGenome Research Institute, USA. We thank the SDS participants and research staffwho made the study possible. Dr. Sanghera and Dr. Howard are the guarantors ofthis work, had full access to all the data, and take full responsibility for theintegrity of the data and the accuracy of data analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
We declare that there is no conflict of interests that could be perceived asprejudicing the impartiality of the research reported.
Electronic supplementary material
12863_2012_1099_MOESM1_ESM.docx
Additional file 1: Figure S1: Exome sequencing reveals the presence of 4 base pair insertion (CCCA)between A and T of ATG start codon in Wnt16a which was confirmed bytargeted sequencing of 30 DNA samples using an ABI 3730 sequencer(Applied Biosystemes Inc. Foster City, USA) and were analyzed usingMutation Surveyor (v4.0.6.). Figure S2. Nusieve-Agarose (3:1) gelshowing wild-type (458 bp), heterozygous insertion(458/462 bp) and homozygous insertion (462 bp) bands inWNt16a gene. Figure S3. Oligonucleotides used toamplify Wnt-16a luciferase reporter constructs. Figure S4.Comparative genomic analysis showing evolution of translation initiationsites in Wnt16a. Arrow indicates the position of insertion in theevolutionarily conserved region at the start codon. (DOCX 393 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Howard, E.W., Been, L.F., Lerner, M. et al. Carriers of a novel frame-shift insertion in WNT16a possess elevatedpancreatic expression of TCF7L2. BMC Genet 14, 28 (2013). https://doi.org/10.1186/1471-2156-14-28
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-14-28