Abstract
Background
The potential adaptive significance of transposable elements (TEs) to the host genomes in which they reside is a topic that has been hotly debated by molecular evolutionists for more than two decades. Recent genomic analyses have demonstrated that TE fragments are associated with functional genes in plants and animals. These findings suggest that TEs may contribute significantly to gene evolution.
Results
We have analyzed two transposable elements associated with genes in the sequenced Drosophila melanogaster y; cn bw sp strain. A fragment of the Antonia long terminal repeat (LTR) retrotransposon is present in the intron of Chitinase 3 (Cht3), a gene located within the constitutive heterochromatin of chromosome 2L. Within the euchromatin of chromosome 2R a full-length Burdock LTR retrotransposon is located immediately 3' to cathD, a gene encoding cathepsin D. We tested for the presence of these two TE/gene associations in strains representing 12 geographically diverse populations of D. melanogaster. While the cathD insertion variant was detected only in the sequenced y; cn bw sp strain, the insertion variant present in the heterochromatic Cht3 gene was found to be fixed throughout twelve D. melanogaster populations and in a D. mauritiana strain suggesting that it maybe of adaptive significance. To further test this hypothesis, we sequenced a 685bp region spanning the LTR fragment in the intron of Cht3 in strains representative of the two sibling species D. melanogaster and D. mauritiana (~2.7 million years divergent). The level of sequence divergence between the two species within this region was significantly lower than expected from the neutral substitution rate and lower than the divergence observed between a randomly selected intron of the Drosophila Alcohol dehydrogenase gene (Adh).
Conclusions
Our results suggest that a 359 bp fragment of an Antonia retrotransposon (complete LTR is 659 bp) located within the intron of the Drosophila melanogaster Cht3 gene is of adaptive evolutionary significance. Our results are consistent with previous suggestions that the presence of TEs in constitutive heterochromatin may be of significance to the expression of heterochromatic genes.
Similar content being viewed by others
Background
The potential adaptive significance of transposable elements (TEs) to the host genomes in which they reside is a topic that has been hotly debated by molecular evolutionists for more than two decades. While the biological importance of TEs seemed self-evident to those scientists involved in their initial discovery [e.g., [1, 2]], the subsequent realization that TEs could be maintained in populations even while imparting slight selective disadvantage to their hosts [e.g., [3–5]] drew into question the presumption of adaptive significance. However, even if TEs can be maintained in populations on a day-to-day basis without providing selective advantage, it does not preclude the possibility that the insertion of TEs in or near genes may, in some instances, be of adaptive advantage.
If TE insertion variants have contributed to adaptive gene evolution, such variants might be expected to be in high frequency or fixed in populations and species. Initial surveys of natural populations of Drosophila melanogaster showing that TE insertion alleles are in uniformly low frequency seemed to negate the adaptive hypothesis [6]. However, the sporadic discovery of degenerate TEs or TE fragments as critical components of functional genes in both plants and animals was sufficient to keep the adaptive hypothesis alive throughout the pre-genomic era [7–11].
The current availability of the complete or nearly complete sequence of select genomes representing a variety of species is providing an unprecedented opportunity to examine the frequency and distribution of TEs in eukaryotic genomes. The results have been dramatic. TEs not only comprise a significant fraction of nearly all eukaryotic genomes thus far sequenced, they have been found to be components of the regulatory and/or coding regions of a surprisingly large number of genes [e.g., [12]]. For example, a recent genomic analysis of 13,799 human genes revealed that approximately 4% harbored retrotransposon sequences within protein-coding regions [13]. Similar results have been recently reported for the nematode Caenorhabditis elegans[14]. Here we analyze the polymorphism of two LTR retrotransposon / host gene associations across geographically widespread D. melanogaster populations and a representative population of the D. melanogaster sibling species, Drosophila mauritiana.
Results
We have initiated a genomic analysis of LTR retrotransposons present in the Drosophila melanogaster genome [e.g., [15]]. Of particular interest is identification of genes harboring TEs and determining if these insertion alleles are in high frequency or fixed among natural populations as would be expected from the adaptive hypothesis. We report here the results of an analysis of two LTR retrotransposon-containing genes located on the second chromosome of the sequenced D. melanogaster y; cn bw sp strain. These two genes present an interesting contrast in that one of them, Chitinase 3 (Cht3), is located within constitutive heterochromatin (Genbank accession: AE002743) while the other, cathD, is located in a euchromatic region of the chromosome (Genbank accession: AE003839). Our findings demonstrate that while the euchromatic cathD insertion variant was not detected in any of the natural populations examined, the insertion variant present in the heterochromatic Cht3 gene was found to be apparently fixed throughout the species. These results are consistent with the view that the presence of TEs in constitutive heterochromatin may have relevance to the expression of heterochromatic genes [e.g., [16, 17]].
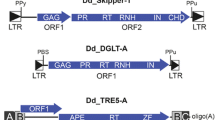
Genomic analysis of the sequenced y; cn bw sp strain of Drosophila melanogaster identified a full-length Burdock LTR retrotransposon located just 3' to the cathD gene and a 359bp LTR fragment (complete LTR is 659 bp) of an Antonia LTR retrotransposon [15] located within an intron of the Cht3 gene (Figure 1). A set of PCR primers were designed to amplify regions of both genes and retrotransposon sequences. Appropriate pairs of gene and element primers were used to detect the presence or absence of the respective retrotransposon inserts associated with each gene in strains representing 12 geographically dispersed populations of D. melanogaster. The results presented in Figure 2 and Table 1 demonstrate that while the Burdock insertion located just 3' to cathD gene is not present in any of 12 strains representing a geographically diverse sampling of natural populations, the Antonia LTR fragment located in the intron of the heterochromatic Cht3 gene is fixed in all 12 strains tested.
Genomic structure of the Cht3 and cathD genes in the Drosophila melanogaster genome. (A) Chromosome 2 illustrating location of Cht3 and cathD genes (red lines) in reference to constitutive heterochromatin (in blue) [34]. Numbers above each red line refer to Flybase cytogenetic placement. (Chromosome not drawn to scale). (B & C) Green arrows represent Flybase-predicted gene regions with corresponding identification. Yellow blocks depict ESTs concordant to the predicted gene region. Blue boxes are predicted exon regions. Red boxes denote LTR position and internal arrows indicate orientation of retroelement. The black line and numbers represent position along the genomic clone sequence which is identified below the figure. Black arrows indicate direction and location of forward (f) or reverse(r) PCR primers. (B) An Antonia LTR fragment (359nt) is inserted in an intron of Cht3 in 12 geographically distinct Drosophila melanogaster strains. (C) A full-length Burdock retroelement, only present in the sequenced y; cn bw sp strain, overlaps the predicted exon boundaries of the cathD gene by 6nt.
It is formally possible that the presence of the Antonia LTR within the Cht3 intron was the result of a chance fixation event prior to the expansion of D. melanogaster around the world. Thus, to further test the adaptive hypothesis we compared the level of sequence divergence within the LTR and its flanking intronic sequence between the two sibling species Drosophila melanogaster and Drosophila mauritiana. If the LTR-containing intron is under stabilizing selection, a lower than neutral rate of substitution would be expected. A total of 685 bp of the Cht3 intron was sequenced. This region spans 264 bp of the 359 bp Antonia LTR fragment. The sequence of this region in a D. melanogaster (Dimonika, Africa) and D. mauritiana (Mauritius, Africa) strain was aligned with the homologous region in the sequenced D. melanogaster y; cn bw sp strain (Figure 3). The two melanogaster strains were 100% identical. The melanogaster sequences were found to be only 1.3% (9 substitutions/685 nucleotide sites) diverged from that of D. mauritiana. This value is significantly less than half of the expected 4.3 % (± 2.7) divergence based on the Drosophila neutral substitution rate of 0.016 (± 0.005) substitutions/site/million year [18] over the estimated 2.7 million years separating the two species [19].
To directly compare the substitution rate for the Cht3 intron with that of another Drosophila gene intron, we randomly selected intron 1 of the Drosophila alcohol dehydrogenase (Adh) gene. Adh is a widely studied Drosophila gene and it has been sequenced in several Drosophila species including D. melanogaster, accession X60793 [20] and D. mauritiana, accession M19264 [21]. The sequence divergence between D. melanogaster and D. mauritiana in the Adh intron 1 (7.9%, Figure 4), is higher than that for the LTR containing Cht3 intron (1.3%). These results strongly suggest that conservative selection has been operating on the LTR containing intron associated with the Drosophila Cht3 gene over the past 2.7 million years.
Discussion
For many years, constitutive heterochromatin was considered to be of little or no functional significance [22]. This view seemed to be supported by early molecular studies showing that heterochromatin consists almost exclusively of highly repeated and middle repetitive DNA [e.g., [23, 24]]. The middle repetitive fraction was viewed as the descendent of once active TEs that had the misfortune of inserting into transcriptionally inert heterochromatin at some point in their evolutionary history [e.g., [6, 20]]. The view of heterochromatin as a genetic wasteland gradually changed with the mapping of a number of functionally important Drosophila genes to constitutive heterochromatin [e.g., [24–31]]. Reexamination of Drosophila constitutive heterochromatin revealed that long stretches of highly repetitive DNA are interrupted by "islands" of retrotransposon sequences [e.g., [32, 33]]. Drosophila genes in heterochromatin are typically associated with these islands of retrotransposons [2, 31, 34–36]. It has been suggested that transposable elements inserted into heterochromatin may locally alter chromatin structure [e.g., [16]]. Our results suggest that in at least some instances, the association of heterochromatic genes with transposable element sequences may be of adaptive significance.
Conclusions
The results presented here are consistent with the hypothesis that a 359 bp fragment of the Antonia retrotransposon located within the intron of the heterochromatic Drosophila melanogaster Cht3 gene may be of adaptive evolutionary significance. Further genomic and molecular analyses will be required to assess the general importance of LTR retrotransposon sequences to the evolution of heterochromatic gene structure and function.
Materials and Methods
Gene Region Annotation
BLASTS of sequenced DNA turned up several instances of genes proximal to an LTR retrotransposon. Sequence retrieval was initiated via BLASTN searches (default parameters- [37]) against the BDGP http://www.fruitfly.org and GenBank http://www.ncbi.nlm.nih.gov databases using LTRs from previously identified Drosophila retroelements as queries [15]. Results with E-values < e-10 were annotated on the corresponding clone, whereupon visual inspection of several annotations confirmed the presence of retroelements proximal to known genes. Selected genes were BLASTed against NCBI's EST database and mapped along with predicted transcript structures from Flybase http://www.flybase.org. Chromosomal location of clones was also determined from Flybase.
PCR
D. melanogaster strains from Dimonika, Niamey, Swaziland, Kenia, Capetown, Cotonake, and India were obtained from Charles F. Aquadro, Cornell University. Germany, Italy, and Antilles strains were obtained from Nikolaj Junakovic, Universitá la Sapienza, Rome, Italy. California and Athens strains are from Daniel Promislow, University of Georgia. D. melanogaster y; cn bw sp strain was obtained from the Bloomington, IN, stock center. The D. mauritiana (241.0) strain was provided by the Bowling Green, OH, Drosophila stock center.
PCR primers were designed with MacVector 7.0 http://www.gcg.com and synthesized by Integrated DNA Technologies (Coralville, LA) (Table 2). Three PCR reactions were performed per strain, per gene. For all PCR reactions, 1.0 μl of a single fly DNA prep [38] was used and amplification was performed in a Hot Top equipped Robocyler Gradient 96 (Stratagene, La Jolla, CA). 10 μl of product was separated on a 1% agarose gel in 0.5× TBE running buffer containing 0.25 μg mL-1 ethidium bromide. Gel images were visualized by UV transillumination.
Cht3 PCR
The PCR products for primer set cht3(f) and cht3(r) and primer set Antonia LTR(f) and Antonia LTR(r) were amplified in a 25 μl reaction containing 3 mM MgCl2, 10X PCR buffer supplied by Pierce (Rockford, IL), 5% DMSO, 0.2 mM dNTPs, 0.5 μM of each primer, and 0.5 U of Taq DNA polymerase supplied by Pierce [Rockford, IL]. The program consisted of an initial incubation at 94°C for 3 min for 1 cycle, a 30 cycle extension at 94°C for 30 sec, 56°C for cht3(f)/cht3(r) primer set or 57°C for Antonia LTR(f)/Antonia LTR(r) primer set for 30 sec, 72°C for 1 min 30 sec, and a 1 cycle final extension of 72°C for 5 min. The PCR products for primer set cht3(f2) and LTR(r) were amplified in a 25 μl reaction containing Expand Long Template PCR System 10X PCR buffer #1 supplied by Roche (Indianapolis, IN), 0.35 mM dNTPs, 0.32 μM of each primer, and 1.3 U of Expand Long Template PCR System DNA polymerase mix supplied by Roche (Indianapolis, IN). The program consisted of an initial incubation at 94°C for 3 min for 1 cycle, a 30 cycle extension at 94°C for 30 sec, 52°C for 30 sec, 68°C for 3 min, and a 1 cycle final extension at 68°C for 5 min.
cathD PCR
The reaction mix and program used for all sets of primers are the same as those described for primer set cht3(f) and cht3(r) and primer set Antonia LTR(f) and Antonia LTR(r) in the Cht3 PCR (above). The annealing temperature for primer set cathD(f) and cathD(r) is 58°C, for primer set Burdock LTR(f) and Burdock element(r) is 59°C, and for primer set cathDff) and Burdock element(r) is 56°C.
Sequencing
PCR products of the Cht3 intron were sequenced in the Molecular Genetics Instrumentation Facility at the University of Georgia. Sequences were aligned with Mac Vector 7.0 and compared to the published y; cn bw sp strain. Substitutions and insertion/deletion sites (indels) were summed for each sequence product and compared to the expected divergence based upon the neutral substitution rate. The expected number of polymorphisms between D. melanogaster and D. mauritiana was calculated based on the Drosophila neutral substitution rate of .016 (± 0.005) substitutions per site/million years [18] on 685 bp over a divergence time of 2.7 million years [19].
PCR analysis testing for the presence of an LTR retroelement feature in two genes, Cht3 and cathD , across three representative Drosophila strains. A negative image is presented for visual clarity. Three PCR reactions were performed per strain, per gene. M = 1 kb ladder, M2 = 0.1 kb ladder. (A) An Antonia LTR fragment is fixed in the intron of the heterochromatic Cht3 gene in all 12 tested strains (only three shown). Cht3-G = cht3 primers (f+r), expected product= 488 bp. L = Antonia LTR primers (f+r), expected product= 272 bp. G2/L = cht3(f2) + Antonia LTR (r) primers, expected product= 3022 bp. (B) A full-length Burdock LTR retrotransposon is found to be associated with cathD only in the sequenced y; cn bw sp strain. cathD – G = cathD primers (f+r), expected product = 461 bp. L = Burdock primers (f+r), expected product = 280 bp. G/L = cathD(f) and Burdock element (r), expected product= 1139 bp.
Nucleotide alignment of a 685 bp Cht3 intron fragment in D.melanogaster and D.mauritiana . Cht3 intron sequence from the Drosophila melanogaster y; cn bw sp strain (accession AE002743). The Antonia LTR stretches from bp 1 – 264, where a black diamond (♦) indicates the end of LTR sequence. Strains representing the D. melanogaster, Africa (Dimonika) population and a strain representing the D. mauritiana, Mauritius population were sequenced. Sequences were aligned using MacVector (See Materials and Methods for details).
Nucleotide alignment of the 659 bp intron 1 of the Adh gene in Drosophila melanogaster and Drosophila mauritiana . Sequences obtained through GenBank for D.melanogaster (accession: X60793, [20] and D.mauritiana (accession: M19264, [21]). Sequences were aligned using MacVector (See Materials and Methods for details).
Note added in proof
The two Cht3 intron fragments descibed in Figure 3 have the following provisional accession numbers in GenBank:
D. melanogaster, Africa - AY081055
D. mauritiana - AY081054
References
McClintock B: Chromosome organization and genic expression. Cold Spr. Harb. Symp. Quant. Biol. 1951, 16: 13-17.
Shapiro J: DNA insertion elements and the evolution of chromosome primary structure. Trends Biochem. Sci. 1977, 2: 622-627. 10.1016/0968-0004(77)90072-X.
Doolittle WF, Sapienza C: Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980, 284: 601-603.
Hickey DA: Selfish DNA: a sexually-transmitted nuclear parasite. Genetics. 1982, 101: 519-531.
Orgel LE, Crick FH: Selfish DNA: the ultimate parasite. Nature. 1980, 284: 604-607.
Charlesworth B, Langley CH: The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989, 23: 251-287. 10.1146/annurev.ge.23.120189.001343.
Britten RJ: DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci USA. 1996, 93: 9374-9377. 10.1073/pnas.93.18.9374.
Britten RJ: Mobile elements inserted in the distant past have taken on important functions. Gene. 1997, 205: 177-182. 10.1016/S0378-1119(97)00399-5.
Brosius J: Retroposons–seeds of evolution. Science. 1991, 251: 753-
Makalowski W, Mitchell GA, Labuda D: Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994, 10: 188-193. 10.1016/0168-9525(94)90254-2.
McDonald JF: Evolution and consequences of transposable elements. Curr Opin Genet Dev. 1993, 3: 855-864.
Brosius J: Genomes were forged by massive bombardments with retroelements and retrosequences. In: Transposable Elements and Genome Evolution. Edited by: McDonald JF. 2000, Dordrecht: Kluwer Academic Publishers, 209-238.
Nekrutenko A, Li WH: Transposable elements are found in a large number of human protein- coding genes. Trends Genet. 2001, 17: 619-621. 10.1016/S0168-9525(01)02445-3.
Ganko EW, Fielman KT, McDonald JF: Evolutionary History of Cer Elements and Their Impact on the C. elegans Genome. Genome Res. 2001, 11: 2066-2074. 10.1101/gr.196201.
Bowen NJ, McDonald JF: Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res. 2001, 11: 1527-1540. 10.1101/gr.164201.
Gatti M, Pimpinelli S: Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992, 26: 239-275. 10.1146/annurev.ge.26.120192.001323.
Spradling AC: Transposable elements and the evolution of heterochromatin. Soc Gen Physiol Ser. 1994, 49: 69-83.
Li WH: Molecular Evolution. Sunderland, MA: Sinauer. 1997
Li YJ, Satta Y, Takahata N: Paleo-demography of the Drosophila melanogaster subgroup: application of the maximum likelihood method. Genes Genet Syst. 1999, 74: 117-127. 10.1266/ggs.74.117.
Laurie CC, Bridgham JT, Choudhary M: Associations between DNA sequence variation and variation in expression of the Adh gene in natural populations of Drosophila melanogaster. Genetics. 1991, 129: 489-499.
Cohn VH, Moore GP: Organization and evolution of the alcohol dehydrogenase gene in Drosophila. Mol Biol Evol. 1988, 5: 154-166.
Hannah A: Localization and function of heterochromatin in Drosophila melanogaster. Adv. Genet. 1951, 4: 87-125.
Appels R, Peacock WJ: The arrangement and evolution of highly repeated (satellite) DNA sequences with special reference to Drosophila. Int Rev Cytol Suppl. 1978, Suppl: 69-126.
Brutlag DL: Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980, 14: 121-144. 10.1146/annurev.ge.14.120180.001005.
Hilliker AJ, Holm DG: Genetic analysis of the proximal region of chromosome 2 of Drosophila melanogaster. I. Detachment products of compound autosomes. Genetics. 1975, 81: 705-721.
Hilliker AJ: Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics. 1976, 83: 765-782.
Ganetzky B: On the components of segregation distortion in Drosophila melanogaster. Genetics. 1977, 86: 321-355.
Pimpinelli S, Sullivan W, Prout M, Sandler L: On biological functions mapping to the heterochromatin of Drosophila melanogaster. Genetics. 1985, 109: 701-724.
Marchant GE: Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. I. Products of compound autosome detachment. Genetics. 1988, 120: 503-517.
Marchant GE: Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. II. Vital loci identified through EMS mutagenesis. Genetics. 1988, 120: 519-532.
Devlin RH, Holm DG, Morin KR, Honda BM: Identifying a single-copy DNA sequence associated with the expression of a heterochromatic gene, the light locus of Drosophila melanogaster. Genome. 1990, 33: 405-415.
Caizzi R, Caggese C, Pimpinelli S: Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics. 1993, 133: 335-345.
Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M: Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci USA. 1995, 92: 3804-3808.
Dimitri P, Junakovic N: Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends Genet. 1999, 15: 123-124. 10.1016/S0168-9525(99)01711-4.
Berghella L, Dimitri P: The heterochromatic rolled gene of Drosophila melanogaster is extensively polytenized and transcriptionally active in the salivary gland chromocenter. Genetics. 1996, 144: 117-125.
Dimitri P, Junokovic N, Arca B: Nested transposons within the intron II of rolled, a heterochromatic gene of Drosophila melanogaster. Dros Res Conf. 1999, 40: 663B-
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR: Type I repressors of P element mobility. Genetics. 1993, 135: 81-95.
Acknowledgements
E.W.G supported through an NIH Genetics Training Grant. This work supported by a National Institutes of Health grant to J.F.M.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
McCollum, A.M., Ganko, E.W., Barrass, P.A. et al. Evidence for the adaptive significance of an LTR retrotransposon sequence in a Drosophila heterochromatic gene. BMC Evol Biol 2, 5 (2002). https://doi.org/10.1186/1471-2148-2-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-2-5