Abstract
Background
The formation of the East African Rift System has decisively influenced the distribution and evolution of tropical Africa’s biota by altering climate conditions, by creating basins for large long-lived lakes, and by affecting the catchment and drainage directions of river systems. However, it remains unclear how rifting affected the biogeographical patterns of freshwater biota through time on a continental scale, which is further complicated by the scarcity of molecular data from the largest African river system, the Congo.
Results
We study these biogeographical patterns using a fossil-calibrated multi-locus phylogeny of the gastropod family Viviparidae. This group allows reconstructing drainage patterns exceptionally well because it disperses very poorly in the absence of existing freshwater connections. Our phylogeny covers localities from major drainage basins of tropical Africa and reveals highly disjunct sister-group relationships between (a) the endemic viviparids of Lake Malawi and populations from the Middle Congo as well as between (b) the Victoria region and the Okavango/Upper Zambezi area.
Conclusions
The current study testifies to repeated disruptions of the distribution of the Viviparidae during the formation of the East African Rift System, and to a central role of the Congo River system for the distribution of the continent’s freshwater fauna during the late Cenozoic. By integrating our results with previous findings on palaeohydrographical connections, we provide a spatially and temporarily explicit model of historical freshwater biogeography in tropical Africa. Finally, we review similarities and differences in patterns of vertebrate and invertebrate dispersal. Amongst others we argue that the closest relatives of present day viviparids in Lake Malawi are living in the Middle Congo River, thus shedding new light on the origin of the endemic fauna of this rift lake.
Similar content being viewed by others
Background
The formation of the East African Rift System (EARS) played a decisive role in the evolution of Africa’s tropical fauna and flora. The climatic, geological, and hydrological consequences of the rifting processes are critical to understand the emergence of the endemic species of the savannahs and the Great Lakes as well as the origin of humankind. Rifting created, disrupted, redirected, and connected major freshwater systems [1].
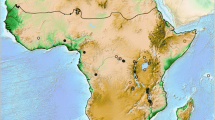
The three largest drainage systems in present-day Central Africa are the Congo-, the Nile-, and the Zambezi system (Figure 1). The Congo River covers an area of ~3.7 million square kilometres and has a length of ~4100 kilometres [2]. It drains part of the EARS (including Lake Tanganyika) and borders to the Nile system in the northeast. The Nile system receives water from Lake Albert and Lake Victoria and drains a region of ~3.2 million square kilometres for almost 7000 kilometres northwards to the Mediterranean Sea. To the south, the Congo system borders to the Zambezi drainage system, which runs ~2600 kilometres, covers an area of 1.3 million square kilometres [2, 3], and includes the southernmost of the Great Lakes, Malawi. The central location of the EARS to these river systems bears witness to the rift’s importance in shaping drainage pattern of tropical Africa, but their history since the Mid-Miocene is only partly resolved and arguably complex [2, 3].
Map of tropical Africa with major drainage systems. The map shows the major drainage systems, framed with bold grey lines, and the sampling points of the study. Lakes and rivers referred to in the text are labelled accordingly; the Upper Zambezi is abbreviated as ‘U. Zambezi’. The dashed outline indicates the maximum expansion of Lake Palaeo-Makgadikgadi, modified after Riedel et al. [4]. Locality details for each sampling point are given in Table 1.
Most suggestions of palaeohydrological connections in tropical Africa are based on circumstantial evidence in combination with general models of rifting and uplifting [2] rather than on geomorphological evidence like in other parts of the continent where palaeochannels can be traced over considerable areas and are reasonably well dated, e.g. [5–7]. The consequential lack of a coherent synthesis or a model of hydrographical connections since the Mid-Miocene severely hampers our understanding of historical biogeography in tropical Africa. This is further complicated by a scarcity of samples from the Congo River system, e.g. [8–11], but see Goodier et al. [12] and Day et al. [13]. The Congo is the largest freshwater system in the centre of the continent and palaeohydrological research indicates that the river and its tributaries have a highly vicissitudinous drainage history. Temporal connections existed to the present day Lake Victoria area [14, 15], Lake Turkana [16, 17], the Upper Zambezi area [18, 19] and the Indian Ocean [19], although the latter connection is controversial [1]. Hence, the analysis of Congolese samples is a prerequisite for our understanding of the evolution and distribution of the freshwater fauna of tropical Africa—including the origins of the endemic radiations in the rift-lakes Malawi and Tanganyika.
Here we study the historical biogeography of the freshwater gastropod family Viviparidae, a taxon widely distributed in the lotic and lentic environments of tropical Africa [20]. This family is of particular interest to studies of historical freshwater biogeography because its dispersal is mostly limited to habitats with existing freshwater connections [21], rendering it an ideal tracer of past and present hydrographic connections. The ovoviviparous and dioecious reproduction mode of viviparids minimizes opportunities for successful vector-mediated migration and a subsequent establishment of a viable population [22, 23]. Accordingly, viviparid fossils are conspicuously absent from African water bodies that were never hydrographically connected to a source, e.g. crater- and oasis lakes [21].
We obtained samples from the major drainage systems in the region, i.e. that of the Nile, the Okavango, the Zambezi as well as the Upper and Middle Congo. Using Bayesian multi-locus phylogenetic analyses and a fossil-calibrated, relaxed molecular clock we studied the diversification and distribution of viviparids in Africa. In light of the impact of the evolution of the EARS on African hydrographic systems and the dependence of our model organism on these systems to disperse, we expect a deep phylogenetic separation between viviparid species east and west of the western branch of the rift system. We furthermore hypothesize this separation to temporarily coincide with the disruption of freshwater migration routes across the evolving rift.
Results
Phylogenetic analyses
The Bayesian analyses of the African Viviparidae yielded one well-supported Neothauma group from Lake Tanganyika (Figure 2; Bayesian posterior probabilities [BPP] 1.0) and two Bellamya clades comprising specimens from the sampled major drainage systems of tropical Africa (Clades I and II in Figure 2; BPP 0.88 and 1.0). Separate analyses of the nuclear and the mitochondrial dataset revealed no phylogenetic incongruences (Additional file 1). Clade I contains four well-supported, reciprocally monophyletic groups of specimens from (i) lakes Albert and Victoria and the Nile River (‘Victoria group’; dark red in Figure 2; BPP 1.0); (ii) the Middle Congo River (‘Congo group’; light green in Figure 2; BPP 1.0); (iii) the rivers Chobe/Cuando and Okavango (‘Okavango group’; light red in Figure 2; BPP 1.0) and (iv) the Malawi Basin (‘Malawi group’; dark green in Figure 2; BPP 1.0). Within Clade I the Congo group is sister to the Malawi group whereas the Victoria group is sister to the Okavango group (both sister-group relationships are supported with BPP 1.0). Clade II consists of a Northern group with specimens from lakes Mweru and Bangweulu (highlighted in blue in Figure 2) and from the Northern Kafue (BPP 0.98) as well as a Southern group with specimens from the Upper Zambezi, the Chobe, and the Southern Kafue (BPP 1.0). Due to lack of support in the two deepest nodes of the phylogeny (BPP < 0.7) we can make no conclusive statement as to the monophyly of either the African Bellamya or the African Viviparidae. These nodes were collapsed into a polytomy in Figure 2. The number of parsimony informative sites and effective sampling sizes are provided in Additional file 1 for each fragment separately.
Multi-locus molecular phylogeny of the African Viviparidae. The phylogeny comprises the genera Bellamya and Neothauma (abbreviated as ‘N.’). The phylogeny was dated using a fossil-calibrated uncorrelated lognormal clock model. Clade I consists of four colour-coded, geographically distinct subgroups from the Middle Congo, Lake Malawi, the Lake Victoria area and the Okavango area. Colours correspond to those in Figure 3. Clade II consists of a Northern group with specimens from lakes Mweru and Bangweulu (highlighted in blue) and from the Northern Kafue as well as a Southern group with specimens from the Upper Zambezi, the Chobe, and the Southern Kafue. Confidence intervals (95%) for age estimates of selected nodes are indicated by grey bars; Bayesian posterior probabilities are given for selected nodes; ‘Out.’ indicates the outgroup. Locality details are provided in Table 1.
The phylogenetic structure revealed by Figure 2 also indicates that morphological identifications and genealogy do not match each other perfectly. A main concern is the systematic position of Bellamya capillata, which has been traditionally reported from many localities in Africa south of the equator [20]. The type locality for this taxon is Lake Malawi, and all other localities from which the species was identified, are in fact inhabited by populations that are deeply divergent from B. capillata in Lake Malawi. Similar issues exist for other Bellamya species (see Discussion). We thus tentatively name specimens with an ambiguous status in current systematics using ‘confer’ (cf.) to emphasize that taxonomic revisions are required.
Molecular clock estimates
We focus on estimates for the ages of the following three nodes in Clade I (Figure 2): Split A—most recent common ancestor (mrca) of the entire clade: 10.47 million years (My) (95% confidence interval [CI]: 6.69-15.04 My); Split B—the split between the Victoria- and the Okavango group: 6.32 My (CI: 3.58-9.96 My); Split C—the split between the Congo- and the Malawi group: 4.52 My (CI: 2.32-7.70 My). The age of the mrca of the Southern group in Clade II is 1.916 My (CI: 0.88-3.31 My). Note that the here estimated mean substitution rate of the COI fragment is 0.93% per My (CI: 0.68-1.18) and therewith very similar to the trait-specific COI Protostomia clock rate for the HKY model suggested by Wilke et al. [24] for dioeceous tropical and subtropical taxa with a generation time of 1 year and a body size of approximately 2–50 mm (mean rate: 1.24% per My; CI: 1.02-1.46). Substitution rates are provided in Additional file 1 for each fragment.
Discussion
The phylogenetic reconstruction reveals two, geographically highly disjunct sister-group relationships in Clade I (Figures 2 and 3-4): the Congo-Malawi-cluster and the Victoria-Okavango-cluster. The groups within each cluster are separated by at least one water divide. This conspicuous spatial pattern stands in sharp contrast to the initially stated expectation that species east of the western branch of the EARS are closer related to each other than to species west of it, and vice versa. Vector-mediated dispersal across water divides is an unlikely explanation for the observed biogeographical pattern for reasons mentioned earlier. Moreover, given the wide spatial overlap of the potential dispersal corridors of the two clusters, we would expect to observe population exchange between taxa from e.g. the Middle Congo and the Nile, or between the Malawi- and the Zambezi/Okavango group, because the involved vectors would have been able to operate in the entire area. Yet the reciprocal monophyletic clades within our phylogeny argue clearly against such an exchange.
Biogeographical model for Bellamya . The model comprises four progressive stages covering roughly (1) Middle Miocene, (2) Late Miocene, (3) Pliocene and (4) Pliocene/Early Pleistocene timeframes. The map shows the current borders and courses of the drainage systems and rivers as well as the current sizes of the lakes. Inset phylogenies track the diversification of Clade I: colours correspond to geographic ranges and the phylogenetic groups in Figure 2. The borders of the suggested ranges are approximations; detailed descriptions of the individual stages are given in the text. Question marks within the dashed areas in stages 3 and 4 indicate potential remnant populations of the clusters from stage 2.
In order to explain our findings we integrated our phylogenetic information with literature data on palaeohydrographical connections to provide a synthesized, spatially and temporarily explicit model of historical freshwater biogeography in tropical Africa. Subsequently, we review recently published studies on African fish biogeography and evaluate similarities and dissimilarities of vertebrate and invertebrate dispersal patterns.
Historical biogeography of Bellamyain tropical Africa
Changes in the structural geology of East Africa associated with uplifting and rifting [25] as well as pronounced moisture cycles over the past 5 My—mainly governed by orbitally-induced insolation at low altitudes [26, 27]—have created a dynamic setting for palaeohydrographical connections and, hence, organismal dispersal. Against this background, we propose the following biogeographical model, which explains the two, spatially disjunct sister-taxon relationships as a consequence of successive changes in hydrographical connections between the Congo River system and its neighbouring water systems (stages 1–4 in Figure 3; refer to Figure 1 for names of water bodies):
-
(1)
The earliest African viviparid fossils were found in tropical regions [15] (Additional file 2). We thus assume that this region is the geographical origin of an ancestral population of Clade I in the Middle/Late Miocene (~12 Ma; Figure 3-1).
-
(2)
Subsequently this ancestral lineage split into the Congo-Malawi- and the Victoria-Okavango-cluster (Split A in Figures 2 and 3-2; mean age ~10.5 My). In the Late Miocene/Early Pliocene the waters of the present day Lake Victoria region drained via a precursor of the Aruwimi River into the Congo [14, 15]. A contemporaneous connection from the Congo system to the headwaters of the Upper Zambezi was suggested via the Kasai River [19, 28, 29], which served as a dispersal route for various fish taxa [11, 12, 30, 31]. This persistent freshwater connection likely also enabled viviparid migration between the present day Lake Victoria region and the Upper Zambezi/Okavango watershed (Figure 3-2). We postulate that contemporaneously the Congo-Malawi-cluster inhabited the Upper Congo catchment including the Chambeshi system—which drained eastward into the Rufiji system until the Pliocene [19]—, the emerging Lake Malawi, and perhaps Palaeolake Rukwa [32] (Figure 3-2). At that time no hydrographical connection existed between the Upper and the Middle Zambezi [28, 33, 34] and there was no faunal exchange between the Victoria-Okavango-cluster and the Congo-Malawi-cluster.
-
(3)
In the Late Miocene/Early Pliocene the Okavango group started to separate from the Victoria group (Split B in Figures 2 and 3-3; mean age ~6.3 My). During a wet phase in East Africa around 3.0-2.8 Ma faunal elements of Congolese origin (e.g. molluscs and sponges) colonised the Turkana Basin for the last time and all subsequent invasions into the basin were of Nilotic origin [16, 17]. This data indicates that the connection of the Victoria region to the Central Congo ceased, at latest with the closure of the Beni Gap in the Early Pleistocene [15] but perhaps earlier [1]; a similar timeframe is assumed for the disruption of the Okavango/Upper Zambezi-Congo connection [3, 28, 35]. We argue that the final separation of the two groups was ultimately governed by these events (Split B in Figure 2) but we acknowledge that the confidence interval for Split B also allows for an earlier separation. Around this time, the ancestors of the current Congo-Malawi groups inhabited an area encompassing the Upper Congo system, the Chambeshi system, and young Lake Malawi, most likely using the northwest corner of the lake’s basin as gateway [3, 19] (Figure 3-3).
-
(4)
Subsequently the connection between Lake Malawi and the Chambeshi/Upper Congo system ceased, which separated the Congolese from the Malawian Bellamya (Split C in Figures 2 and 3-4; mean age ~4.5 My). This faunal disruption may have been a direct consequence of rifting in the Mweru-Mweru Wantipa fault zone and in the northern part of the Malawi Rift [36–38]. The extant viviparid fauna of the lakes Mweru and Bangweulu is however no remnant of this former Congo-Malawi-cluster (Figure 2). In fact, age and phylogenetic position of the Mweru/Bangweulu viviparids suggest a re-colonization of the two lakes from the Zambezi via the Kafue River [39], perhaps after a climatically-induced Pleistocene ecosystem crisis comparable to those in nearby Lake Malawi in the east [40–42] and Lake Palaeo-Makgadikgadi to the west [43]. Moreover, the polyphyletic pattern of the Mweru/Bangweulu viviparids indicates that the two lakes may have been colonized multiple times from the Zambezi River system (Figures 2 and 3-4). In the Late Pliocene/Early Pleistocene the Upper Zambezi was captured by the Middle Zambezi [28, 34], which enabled secondary contact between the Okavango group and Clade II (Figures 2 and 3-4). The observable geographical overlap between the two groups coincides with the area of Lake Palaeo-Makgadikgadi, which existed in consecutive phases over the Middle and Late Pleistocene [43–45] (Figures 1 and 3-4). This lake may have stirred populations of initially different geographic origin (e.g. the Okavango group and Clade II) and, hence, superimposed previous spatial patterns resulting in the current overlapping distributions of Bellamya groups.
Lake Malawi
The phylogeny presented here reveals an unanticipated sister relationship of Lake Malawi’s extant Bellamya fauna to that of the Middle Congo. In light of the well-supported reciprocal monophyly of both groups we argue that the separation of the Congo-Malawi-cluster (Split C in Figures 2 and 3-4) was a consequence of the disruption of hydrological connections and therewith migration routes (e.g. due to rifting processes) rather than of dispersal from the Congo River system into Lake Malawi or vice versa. Dispersal would most likely have resulted in a pattern where the sink population is nested within the paraphyletic source population instead of the observed reciprocal and strict regional monophyly. The Bellamya of the lakes Mweru and Bangweulu, which re-colonized these water bodies from the Zambezi system, illustrate such a case of dispersal (Figure 2). We hence also conclude that the general dominance of regionally confined monophyletic clusters in our dataset points toward a prevalence of vicariance over dispersal processes in the historical biogeography of the taxon—in contrast to, e.g., some fish taxa (see below).
Viviparid fossils with an age of up to four million years are reported from the Chiwondo Beds in the Northwest of the Malawi Basin [46]. Because the mean age of the split between the Malawi and the Congo group is of a similar age, our results do neither reject the possibility of an uninterrupted evolutionary Bellamya lineage in Lake Malawi (the founder-flush model in Schultheiß et al. [47, 48]) nor the possibility that the lake was re-colonized after its Pliocene fauna went extinct, e.g. during severe lake level low stands [41]. In any case, the onset of molecular diversification of the modern endemic Malawi group started considerably later (1.39 Ma; CI: 0.56-2.61 My) than the deposition of the Plio-Pleistocene Chiwondo Bed fossils. Thus it has paralleled the diversification of the endemic Lanistes taxa of Lake Malawi [48].
Lake Tanganyika
Although Neothauma is currently endemic to Lake Tanganyika, this has not been the case from the Late Miocene to the Early Pleistocene when the genus radiated in Palaeolake Obweruka [15]. This palaeolake occupied the Lake Albert-Edward region and was similar in size and depth to Lake Tanganyika prior to the uplift of the Rwenzori Mountains ~2.3 Ma [14, 49]. The molecular phylogeny indicates that the Neothauma lineage from Lake Tanganyika evolved fairly isolated from the African Bellamya (Figure 2). A similar spatial, temporal, and genetic distinctness is reported for some fish taxa of Lake Tanganyika [31, 50, 51]. It appears reasonable to assume that the lake served as a refuge and harbours the sole survivors of the formerly more widely distributed and morphologically diverse genus Neothauma. Moreover, the isolated evolutionary history of Lake Tanganyika’s viviparid fauna implies that for the timeframe and migration events discussed in this paper the lake served neither as a gateway nor as a dispersal route.
Comparative biogeography of afrotropical freshwater taxa
This paper is to our knowledge the first invertebrate study investigating patterns of historical freshwater biogeography in tropical Africa using molecular data. Against the background of recent studies on African fish biogeography we here review patterns of vertebrate and invertebrate biogeography with a strong focus on dispersal abilities, hybridization, and migration routes.
A striking difference between viviparid and fish patterns lies in the phylogenetic relationships of taxa from the modern Lake Victoria region: fish specimens from the Lake Victoria region cluster generally with other East African species, e.g. [31, 50, 52, 53]. In contrast, the sister-taxon relationship between viviparids from the Lake Victoria region and those from the Okavango/Upper Zambezi is to date unique among the afrotropical freshwater fauna. This pattern is remarkable because connecting these two drainage systems requires crossing two divides: (A) the Congo/Upper Zambezi water divide via a precursor of the Kasai River and (B) crossing the emerging western branch of the EARS. To provide a hypothesis as to the cause of the difference of biogeographical patterns between fish and viviparid taxa, we suggest that the rifting process was the vicariance event that caused the main phylogenetic separation of fish taxa into species east and west of the western branch of the EARS. At this stage, however, the afrotropical Viviparidae were already separated into the Congo-Malawi cluster and the Victoria-Okavango cluster (in the aftermath of Split A; Figures 2 and 3). We argue that this initial Split A was invoked by a migration barrier—perhaps in the Congo Basin—that inhibited viviparid dispersal yet allowed fish dispersal. Intrinsic biological properties may create differential abilities to overcome ecological, behavioural, and physical barriers [30], e.g. differences in mobility may cause differential migration through corridors which only exit during annual flooding. For example, two major flooding events since 2010 promoted the migration of fish from the Okavango Delta into the formerly dry Boteti river bed. However, Bellamya—which lives in permanent overspills of the Okavango Delta such as the Thamalakane River—did not migrate into the Boteti River (FR pers. observation, 2010, 2011, and 2013).
Eventually, the rifting process separated the two viviparid clusters into the four modern groups (i.e. the Congo-, Malawi, Okavango-, and Victoria group; Splits B and C) subsequently to Split A. Each of these groups is (much like in fish) confined to one side of the western branch of the EARS.
This pattern and comparative biogeography sheds new light on freshwater migration corridors over the emerging rift. African tigerfish, for example, were suggested to have migrated during the Late Miocene along an east–west corridor from the Tanzanian Rufiji-Ruaha system via the Luangwa to the Upper Zambezi and the Okavango [12], crossing the young rift at the narrow Rukwa-Rungwe region prior to the uplift of the Ufipa plateau [54]. A similar corridor over the rift appears to have been feasible for mastacembelid eels [51] and, in fact, also for viviparids: The Congo-Malawi-cluster would likewise have occupied the Rukwa-Rungwe region (green area in Figure 3-2). It would hence be highly improbable that members of the Victoria-Okavango cluster migrated contemporaneously from the Victoria region through this corridor across the Rukwa-Rungwe area into the Upper Zambezi/Okavango. This would imply that members of this cluster migrated along the same hydrographical system as the members of the Congo-Malawi-cluster, yet without disrupting the here revealed biogeographical pattern. Consequently, we suggest the existence of a second east–west migration corridor (red area in Figure 3-2) connecting the Victoria region with the Okavango/Upper Zambezi area.
With the exception of some branches in Clade II, our phylogeny generally features strict regional monophyly—a pattern often absent in biogeographical studies of fish in tropical Africa, e.g. [11, 12, 55]. This absence is generally attributed to hybridization and active, long distance dispersal, e.g. [8, 11, 12, 31, 52], which hence appear to have played only a minor role in the historical biogeography of viviparids. Therewith, superimposition of underlying distributional patterns by recent active dispersal or hybridization seems negligible for this gastropod group, which in turn facilitates the reconstruction of original migration corridors.
The decisive phylogenetic pattern notwithstanding, further sampling is required to substantiate the proposed model. In particular, more sampling from the central Congo area is desirable despite logistic challenges, e.g., to search for putative remnants of the Victoria-Okavango-cluster (red dashed area in Figure 3-3). Furthermore, specimens are required from the Rufiji-Ruaha system to test our hypotheses associated with the Malawi-Congo-cluster (green dashed area in Figure 3-3). Eventually, more comprehensive sampling of the Lower Zambezi may shed light on the origins and evolution of Clade II. It is noteworthy that such an extension of the dataset will also enable a systematic revision of the genus Bellamya, as our work reported considerable incongruences between traditional taxonomy and the results of molecular phylogenetic analyses, e.g. [47, 56].
Conclusions
We have presented a first integrated model of historic freshwater invertebrate biogeography in tropical Africa. By comparing our results to fish data we demonstrate the importance of broad geographical and taxonomic sampling encompassing taxa with different dispersal abilities and life history traits. Our analysis shows that the biographical history of viviparids in tropical Africa is driven by consecutive vicariance events rather than by dispersal and is closely linked to the formation of the EARS. Moreover, we demonstrate that the Congo drainage system plays a central role in the distribution of the continent’s freshwater fauna during the late Cenozoic. Our data also sheds new light on the geographic origin of the endemic fauna of Lake Malawi and reveals its close phylogenetic relationship to Congolese populations. We hope that future work, with a further inclusion of invertebrate taxa, will help to refine our understanding of biogeographical patterns of tropical Africa’s freshwater fauna.
Methods
Sampling and species identification
Specimens of Viviparidae were collected between 2006 and 2012 in tropical Africa (Figure 1; Table 1). There are two extant genera of the family on the continent: the widely distributed Bellamya and the monospecific Neothauma, which is endemic to Lake Tanganyika [20]. Due to the ambiguous phylogenetic relationship between the two genera [47, 56], we furthermore added the following closely related Asian viviparid genera to our dataset: Anulotaia, Idiopoma, and Trochotaia. Material was identified before molecular analysis based on shell morphology following the currently accepted taxonomy of African Viviparidae, e.g. [20].
DNA extraction and amplification
We used a CTAB protocol for DNA extraction [57] and amplified four DNA fragments: the cytochrome oxidase subunit I (COI) with universal [58] and specific primers [47], the mitochondrial large subunit of ribosomal RNA (LSU rRNA) [59], a fragment of the nuclear LSU rRNA [60], and the nuclear histone 3 [61]. PCR conditions were as given in [62] and sequencing was performed on an ABI3730XL sequencer (LGC Genomics Germany, Berlin). Sequences of all fragments were aligned unambiguously with ClustalW 1.4 [63] in BioEdit 7.1 [64] and checked by eye.
Phylogenetic analyses and molecular clock estimations
We ran five independent Bayesian phylogenetic reconstructions (partitioned into the four DNA fragments) in BEAST 1.75 [65] to ensure convergence on the same posterior probability distribution. An unlinked HKY + Γ substitution model was utilized because more complex models showed indications of overfitting in preliminary runs; as tree prior we used a Yule model. In order to estimate divergence times we employed a fossil-based calibration using the earliest bellamyinid viviparid from Africa and the oldest Neothauma fossil (for an in-depth discussion see Additional file 2). The oldest Bellamya occurs in the Iriri Member of the Napak Formation at Napak and is ~19 My old [66]. To implement this calibration point, we used a uniform prior over the root of the phylogeny spanning 23–13 My to account for dating uncertainty and the various published hypotheses as to when the Bellamyinae invaded Africa from Asia, e.g. [66–68]. The oldest Neothauma fossil (Neothauma hattinghi Van Damme & Pickford, 1999) is reported from the Upper Miocene Kakara Formation from the western branch of the East African Rift System and is 10–11 My old [15, 69]. For this second calibration point we used a normally distributed age prior (mean age: 10.5 My, standard deviation: 0.5 My) over the basal node leading to the monophyletic Neothauma clade (i.e. stem calibration in BEAST 1.75). Uncorrelated lognormal relaxed clock models were chosen to account for lineage specific rate heterogeneity within each partition; mixing of the Markov chains was monitored with Tracer 1.5 [70].
Ancestral area/state reconstructions as implemented in, e.g., Lagrange, BEAST and Mesquite are not feasible with the present datasets for two reasons. First, the basal topology of the African viviparids is unresolved, whereas a dichotomy is needed to reconstruct the ancestral state and the dichotomy-enforcing algorithm of BEAST would result in low node supports. Secondly, the four major groups of Clade I (Malawi, Congo, Okavango, and Victoria group) form a dichotomous topology (Figure 2), resulting in non-informative ancestral state reconstructions for Split C (Malawi/Congo) and Split B (Okavango/Victoria), and an ambiguous state reconstruction for the most basal Split A due to lacking information (unresolved topology) of the most common recent ancestor.
Availability of supporting data
The data sets supporting the results of this article are available in the treeBASE repository, http://purl.org/phylo/treebase/phylows/study/TB2:S15381.
Accession numbers
All DNA sequences used in this study are available from NCBI GenBank. Accession numbers are provided in Table 1.
Abbreviations
- EARS:
-
East African Rift System
- BPP:
-
Bayesian posterior probabilities
- cf.:
-
Confer
- mrca:
-
Most recent common ancestor
- My:
-
Million years
- CI:
-
Confidence interval
- COI:
-
Cytochrome oxidase subunit I
- LSU rRNA:
-
Large subunit of ribosomal RNA.
References
Roberts EM, Stevens NJ, O’Connor PM, Dirks PHGM, Gottfried MD, Clyde WC, Armstrong RA, Kemp AIS, Hemming S: Initiation of the western branch of the East African Rift coeval with the eastern branch. Nat Geosci. 2012, 5: 289-294. 10.1038/ngeo1432.
Goudie AS: The drainage of Africa since the Cretaceous. Geomorphology. 2005, 67: 437-456. 10.1016/j.geomorph.2004.11.008.
Beadle LC: The Inland Waters of Tropical Africa. 1981, London and New York: Longman
Riedel F, von Rintelen T, Erhardt S, Kossler A: A fossil Potadoma (Gastropoda: Pachychilidae) from Pleistocene central Kalahari fluvio-lacustrine sediments. Hydrobiologia. 2009, 636: 493-498. 10.1007/s10750-009-9958-7.
Lihoreau F, Boisserie J-R, Viriot L, Coppens Y, Likius A, Mackaye HT, Tafforeau P, Vignaud P, Brunet M: Anthracothere dental anatomy reveals a late Miocene Chado-Libyan bioprovince. Proc Natl Acad Sci USA. 2006, 103: 8763-8767. 10.1073/pnas.0603126103.
Griffin DL: Aridity and humidity: two aspects of the late Miocene climate of North Africa and the Mediterranean. Paleogeogr Paleoclimatol Paleoecol. 2002, 182: 65-91. 10.1016/S0031-0182(01)00453-9.
Griffin DL: The late Neogene Sahabi rivers of the Sahara and their climatic and environmental implications for the Chad Basin. J Geol Soc. 2006, 163: 905-921. 10.1144/0016-76492005-049.
Joyce DA, Lunt DH, Bills R, Turner GF, Katongo C, Duftner N, Sturmbauer C, Seehausen O: An extant cichlid fish radiation emerged in an extinct Pleistocene lake. Nature. 2005, 435: 90-95. 10.1038/nature03489.
Wilson AB, Glaubrecht M, Meyer A: Ancient lakes as evolutionary reservoirs: evidence from the thalassoid gastropods of Lake Tanganyika. Proc R Soc Lond B. 2004, 271: 529-536. 10.1098/rspb.2003.2624.
Schultheiß R, Ndeo OW, Malikwisha M, Marek C, Bößneck U, Albrecht C: Freshwater molluscs of the Eastern Congo: notes on taxonomy, biogeography and conservation. Afr Invertebr. 2011, 52: 265-284. 10.5733/afin.052.0204.
Day JJ, Bills R, Friel JP: Lacustrine radiations in African Synodontis catfish. J Evolution Biol. 2009, 22: 805-817. 10.1111/j.1420-9101.2009.01691.x.
Goodier SAM, Cotterill FPD, O’Ryan C, Skelton PH, de Wit MJ: Cryptic diversity of African tigerfish (genus Hydrocynus) reveals palaeogeographic signatures of linked Neogene geotectonic events. PLoS One. 2011, 6: e28775-10.1371/journal.pone.0028775.
Day JJ, Peart CR, Brown KJ, Friel JP, Bills R, Moritz T: Continental diversification of an African catfish radiation (Mochokidae: Synodontis). Syst Biol. 2013, 62: 351-365. 10.1093/sysbio/syt001.
Pickford M, Senut B, Hadoto D: Geology and Paleobiology of the Albertine Rift Valley, Uganda-Zaire. Volume I: Geology. 1993, Orleans: Centre international pour la formation et les échanges geologiques, [Publication Occasionnelle]
Van Damme D, Pickford M: The late Cenozoic Viviparidae (Mollusca, Gastropoda) of the Albertine Rift Valley (Uganda-Congo). Hydrobiologia. 1999, 390: 171-217.
Van Bocxlaer B, Van Damme D, Feibel CS: Gradual versus punctuated equilibrium evolution in the Turkana Basin molluscs: evolutionary events or biological invasions?. Evolution. 2008, 62: 511-520. 10.1111/j.1558-5646.2007.00296.x.
Williamson PG: Evidence for an early Plio-Pleistocene rainforest expansion in East Africa. Nature. 1985, 315: 487-489. 10.1038/315487a0.
Moore AE, Larkin PA: Drainage evolution in south-central Africa since the breakup of Gondwana. S Afr J Geol. 2001, 104: 47-68. 10.2113/104.1.47.
Stankiewicz J, de Wit MJ: A proposed drainage evolution model for Central Africa—Did the Congo flow east?. J Afr Earth Sci. 2006, 44: 75-84. 10.1016/j.jafrearsci.2005.11.008.
Brown DS: Freshwater Snails of Africa and Their Medical Importance. 1994, London: Taylor & Francis
Van Bocxlaer B, Verschuren D, Schettler G, Kröpelin S: Modern and early Holocene mollusc fauna of the Ounianga lakes (northern Chad): implications for the palaeohydrology of the central Sahara. J Quaternary Sci. 2011, 26: 433-447. 10.1002/jqs.1469.
Kappes H, Haase P: Slow, but steady: dispersal of freshwater molluscs. Aquat Sci Res Across Bound. 2012, 74: 1-14. 10.1007/s00027-011-0187-6.
Van Damme D: The Freshwater Mollusca of Northern Africa: Distribution, Biogeography, and Palaeoecology. Volume 25. 1984, Dordrecht, The Netherlands: W. Junk, [Developments in Hydrobiology]
Wilke T, Schultheiß R, Albrecht C: As time goes by: a simple fool’s guide to molecular clock approaches in invertebrates. Am Malacol Bull. 2009, 27: 25-45. 10.4003/006.027.0203.
Chorowicz J: The East African rift system. J Afr Earth Sci. 2005, 43: 379-410. 10.1016/j.jafrearsci.2005.07.019.
Trauth MH, Maslin MA, Deino A, Strecker MR: Late Cenozoic moisture history of East Africa. Science. 2005, 309: 2051-2053. 10.1126/science.1112964.
Trauth MH, Larrasoaña JC, Mudelsee M: Trends, rhythms and events in Plio-Pleistocene African climate. Quaternary Sci Rev. 2009, 28: 399-411. 10.1016/j.quascirev.2008.11.003.
Moore AE, Cotterill FPD, Main MPL, Williams HB: The Zambezi River. Large Rivers. Edited by: Gupta A. 2007, Chichester, UK: John Wiley & Sons, Ltd, 311-332.
Veatch AC: Evolution of the Congo Basin. 1935, New York: The Geological Society of America, [Memoirs of the Geological Society of America, vol. 3]
Bell-Cross G: The distribution of fishes in Central Africa. Fish Res Bull Zambia. 1966, 4: 3-20.
Schwarzer J, Swartz ER, Vreven E, Snoeks J, Cotterill FPD, Misof B, Schliewen UK: Repeated trans-watershed hybridization among haplochromine cichlids (Cichlidae) was triggered by Neogene landscape evolution. Proc R Soc Lond B. 2012, 279: 4389-4398. 10.1098/rspb.2012.1667.
Cohen AS, Van Bocxlaer B, Todd JA, McGlue M, Michel E, Nkotagu HH, Grove AT, Delvaux D: Quaternary ostracodes and molluscs from the Rukwa Basin (Tanzania) and their evolutionary and paleobiogeographic implications. Palaeogeogr Palaeoclimatol Palaeoecol. 2013, 392: 79-97.
Thomas DSG, Shaw PA: The Kalahari Environment. 1991, Cambridge, UK: Cambridge University Press
Moore AE, Cotterill FPD, Eckardt FD: The evolution and ages of Makgadikgadi Palaeo-Lakes: consilient evidence from Kalahari drainage evolution, Botswana. S Afr J Geol. 2012, 115: 385-413. 10.2113/gssajg.115.3.385.
Bell-Cross G: Physical barriers separating the fishes of the Kafue and middle Zambezi river systems. Fish Res Bull Zambia. 1965, 4: 97-98.
Mondeguer A, Ravenne C, Masse P, Tiercelin J-J: Sedimentary basins in an extension and strike-slip background: the “South Tanganyika troughs complex”, East African Rift. Bull Soc géol France. 1989, 5: 501-522.
Tiercelin J-J, Lezzar K-E: A 300 Million Years History of Rift Lakes in Central and East Africa: An Updated Broad Review. The East African Great Lakes: Limnology, Palaeolimnology and Biodiversity. Volume 12. Edited by: Odada E, Olago D. 2004, Dordrecht, The Netherlands: Springer Netherlands, 3-60. [Advances in Global Change Research]
Banks NL, Bardwell KA, Musiwa S: Karoo Rift Basins of the Luangwa Valley, Zambia. Hydrocarbon Habitat in Rift Basins. Edited by: Lambiase JJ. 1995, London: Geological Society, 285-295. [Special Publication, vol. 80]
Cotterill FPD, de Wit MJ: Geoecodynamics and the Kalahari epeirogeny: linking its genomic record, tree of life and palimpsest into a unified narrative of landscape evolution. S Afr J Geol. 2011, 114: 489-514. 10.2113/gssajg.114.3-4.489.
Delvaux D: Age of Lake Malawi (Nyasa) and water level fluctuations. Mus roy Afr centr, Tervuren (Belg), Dept Geol Min, Rapp ann 1993 & 1994. 1995, 1995: 99-108.
Scholz CA, Johnson TC, Cohen AS, King JW, Peck JA, Overpeck JT, Talbot MR, Brown ET, Kalindekafe L, Amoako PYO, Lyons RP, Shanahan TM, Castañeda IS, Heil CW, Forman SL, McHargue LR, Beuning KR, Gomez J, Pierson J: East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc Natl Acad Sci USA. 2007, 104: 16416-16421. 10.1073/pnas.0703874104.
Cohen AS, Stone JR, Beuning KRM, Park LE, Reinthal PN, Dettman D, Scholz CA, Johnson TC, King JW, Talbot MR, Brown ET, Ivory SJ: Ecological consequences of early Late Pleistocene megadroughts in tropical Africa. Proc Natl Acad Sci USA. 2007, 104: 16422-16427. 10.1073/pnas.0703873104.
Burrough SL, Thomas DSG, Bailey RM: Mega-Lake in the Kalahari: A Late Pleistocene record of the Palaeolake Makgadikgadi system. Quaternary Sci Rev. 2009, 28: 1392-1411. 10.1016/j.quascirev.2009.02.007.
Haddon IG, McCarthy TS: The Mesozoic–Cenozoic interior sag basins of Central Africa: The Late-Cretaceous–Cenozoic Kalahari and Okavango basins. J Afr Earth Sci. 2005, 43: 316-333. 10.1016/j.jafrearsci.2005.07.008.
McFarlane MJ, Eckardt FD: Lake Deception: a new Makgadikgadi palaeolake. Botsw Notes Rec. 2008, 38: 195-201.
Schrenk F, Bromage TG, Gorthner A, Sandrock O: Paleoecology of the Malawi Rift: vertebrate and invertebrate faunal contexts of the Chiwondo beds, northern Malawi. J Hum Evol. 1995, 28: 59-70. 10.1006/jhev.1995.1006.
Schultheiß R, Wilke T, Jørgensen A, Albrecht C: The birth of an endemic species flock: demographic history of the Bellamya group (Gastropoda, Viviparidae) in Lake Malawi. Biol J Linnean Soc. 2011, 102: 130-143. 10.1111/j.1095-8312.2010.01574.x.
Schultheiß R, Van Bocxlaer B, Wilke T, Albrecht C: Old fossils—young species: evolutionary history of an endemic gastropod assemblage in Lake Malawi. Proc R Soc Lond B. 2009, 276: 2837-2846. 10.1098/rspb.2009.0467.
Ring U: Extreme uplift of the Rwenzori Mountains in the East African Rift, Uganda: Structural framework and possible role of glaciations. Tectonics. 2008, 27: TC4018-
Day JJ, Wilkinson M: On the origin of the Synodontis catfish species flock from Lake Tanganyika. Biol Lett. 2006, 2: 548-552. 10.1098/rsbl.2006.0532.
Brown K, Rüber L, Bills R, Day J: Mastacembelid eels support Lake Tanganyika as an evolutionary hotspot of diversification. BMC Evol Biol. 2010, 10: 188-10.1186/1471-2148-10-188.
Koblmüller S, Sturmbauer C, Verheyen E, Meyer A, Salzburger W: Mitochondrial phylogeny and phylogeography of East African squeaker catfishes (Siluriformes: Synodontis). BMC Evol Biol. 2006, 6: 49-10.1186/1471-2148-6-49.
Pinton A, Agnèse J-F, Paugy D, Otero O: A large-scale phylogeny of Synodontis (Mochokidae, Siluriformes) reveals the influence of geological events on continental diversity during the Cenozoic. Mol Phylogenet Evol. 2013, 66: 1027-1040. 10.1016/j.ympev.2012.12.009.
Delvaux D, Kervyn F, Macheyeki AS, Temu EB: Geodynamic significance of the TRM segment in the East African Rift (W-Tanzania): Active tectonics and paleostress in the Ufipa plateau and Rukwa basin. J Struct Geol. 2012, 37: 161-180.
Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, Turner GF: Age of cichlids: new dates for ancient lake fish radiations. Mol Biol Evol. 2007, 24: 1269-1282. 10.1093/molbev/msm050.
Sengupta ME, Kristensen TK, Madsen H, Jørgensen A: Molecular phylogenetic investigations of the Viviparidae (Gastropoda: Caenogastropoda) in the lakes of the Rift Valley area of Africa. Mol Phylogenet Evol. 2009, 52: 797-805. 10.1016/j.ympev.2009.05.007.
Wilke T, Davis GM, Qiu DC, Spear RC: Extreme mitochondrial sequence diversity in the intermediate schistosomiasis host Oncomelania hupensis robertsoni: Another case of ancestral polymorphism?. Malacologia. 2006, 48: 143-157.
Folmer O, Black M, Hoeh WR, Lutz R, Vrijenhoek R: DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994, 3: 294-299.
Palumbi S, Martin A, Romano S, Mcmillian WO, Stice L, Grabowski G: The Simple Fool’s Guide to PCR. 1991, Honolulu: University of Hawaii
Park JK, Foighil DÓ: Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Mol Phylogenet Evol. 2000, 14: 75-88. 10.1006/mpev.1999.0691.
Colgan DJ, Ponder WF, Eggler PE: Gastropod evolutionary rates and phylogenetic relationships assessed using partial 28S rDNA and histone H3 sequences. Zool Scr. 2000, 29: 29-63. 10.1046/j.1463-6409.2000.00021.x.
Schultheiß R, Albrecht C, Bößneck U, Wilke T: The neglected side of speciation in ancient lakes: phylogeography of an inconspicuous mollusc taxon in lakes Ohrid and Prespa. Hydrobiologia. 2008, 615: 141-156. 10.1007/s10750-008-9553-3.
Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994, 22: 4673-4680. 10.1093/nar/22.22.4673.
Hall TA: BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999, 41: 95-98.
Drummond AJ, Suchard MA, Xie D, Rambaut A: Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012, 29: 1969-1973. 10.1093/molbev/mss075.
Pickford M: Palaeoenvironments of Early Miocene hominoid-bearing deposits at Napak, Uganda, based on terrestrial molluscs. Annales de Paléontologie. 2004, 90: 1-12.
Kat PW: Biogeography and evolution of African fresh-water mollusks – implications of a Miocene assemblage from Rusinga-Island, Kenya. Palaeontology. 1987, 30: 733-742.
Van Damme D, Van Bocxlaer B: Freshwater Molluscs of the Nile Basin, Past and Present. The Nile – Origin, Environments, Limnology and Human Use. Edited by: Dumont HJ. 2009, Dordrecht, The Netherlands: Springer Netherlands
Van Damme D, Pickford M: The late Cenozoic Thiaridae (Mollusca, Gastropoda, Cerithioidea) of the Albertine Rift Valley (Uganda-Congo) and their bearing on the origin and evolution of the Tanganyikan thalassoid malacofauna. Hydrobiologia. 2003, 498: 1-83. 10.1023/A:1026298512117.
Rambaut A, Drummond AJ: Tracer 1.5. 2009, Available from http://beast.bio.ed.ac.uk/Tracer
Acknowledgements
We are grateful to Andrew Goudie, Jacek Stankiewicz, and Woody Cotterill for an evaluation of the model. Daniel Engelhard, Thies Geertz, Andreas Schumann, Jean-Papy Mongindo, Ernest Tambwe, Martin Pickford, Ezra Mussime and Kitam Ali are thanked for support in the field and access to museum specimens. Romy Richter, Catharina Clewing, and Silvia Nachtigall provided assistance in the laboratory. The manuscript benefited from careful readings by Woody Cotterill, Thomas Wilke, Gene Hunt, and two anonymous reviewers. The study was funded by the DFG projects AL 1076/5-2, AL 1076/6-2, RI 1738/3-1, and RI 809/20-1,2. BVB is a Peter-Buck Fellow of the Smithsonian Institution; his fieldwork related to the present study was funded by the Flanders Research Foundation and the Boyekoli Ebale Congo 2010 expedition through the Belgian Federal Directorate General for Development Cooperation (DGDC) and the Belgian Science Policy (BELSPO).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RS analysed the data, developed the model and led the writing of the manuscript. BVB developed the fossil framework of the study and contributed to the model building and writing. FR, TvR, and CA designed and coordinated the study; TvR and CA provided the DNA sequences. Material was collected by FR, BVB, and RS. All authors read and approved the final manuscript.
Electronic supplementary material
12862_2013_2737_MOESM1_ESM.pdf
Additional file 1: The supplementary file contains additional information on the phylogenetic analyses conducted during this study. We furthermore provide the results of the separate analyses of nuclear and mitochondrial data. (PDF 272 KB)
12862_2013_2737_MOESM2_ESM.pdf
Additional file 2: The supplementary file contains additional information on the fossil record of the African Viviparidae and the dating constrains employed in the phylogenetic analysis of this study.(PDF 250 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Schultheiß, R., Van Bocxlaer, B., Riedel, F. et al. Disjunct distributions of freshwater snails testify to a central role of the Congo system in shaping biogeographical patterns in Africa. BMC Evol Biol 14, 42 (2014). https://doi.org/10.1186/1471-2148-14-42
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-14-42