Abstract
Background
Morphostasis of traits in different species is necessary for reconstructing the evolutionary history of complex characters. Studies that place these species into a molecular phylogenetic context test hypotheses about the transitional stages that link divergent character states. For instance, the transition from a phagotrophic mode of nutrition to a phototrophic lifestyle has occurred several times independently across the tree of eukaryotes; one of these events took place within the Euglenida, a large group of flagellates with diverse modes of nutrition. Phototrophic euglenids form a clade that is nested within lineages of phagotrophic euglenids and that originated through a secondary endosymbiosis with green algae. Although it is clear that phototrophic euglenids evolved from phagotrophic ancestors, the morphological disparity between species representing these different nutritional modes remains substantial.
Results
We cultivated a novel marine euglenid, Rapaza viridis n. gen. et sp. ("green grasper"), and a green alga, Tetraselmis sp., from the same environment. Cells of R. viridis were comprehensively characterized with light microscopy, SEM, TEM, and molecular phylogenetic analysis of small subunit rDNA sequences. Ultrastructural and behavioral observations demonstrated that this isolate habitually consumes a specific strain of Tetraselmis prey cells and possesses a functional chloroplast that is homologous with other phototrophic euglenids. A novel feeding apparatus consisting of a reduced rod of microtubules facilitated this first and only example of mixotrophy among euglenids. R. viridis also possessed a robust photoreception apparatus, two flagella of unequal length, euglenoid movement, and a pellicle consisting of 16 strips and one (square-shaped) whorl of posterior strip reduction. The molecular phylogenetic data demonstrated that R. viridis branches as the nearest sister lineage to phototrophic euglenids.
Conclusions
The unusual combination of features in R. viridis combined with its molecular phylogenetic position completely conforms to the expected transitional stage that occurred during the early evolution of phototrophic euglenids from phagotrophic ancestors. The marine mixotrophic mode of nutrition, the preference for green algal prey cells, the structure of the feeding apparatus, and the organization of the pellicle are outstanding examples of morphostasis that clarify pivotal stages in the evolutionary history of this diverse group of microbial eukaryotes.
Similar content being viewed by others
Background
Morphostasis and transitional traits
The reconstruction of early stages in the evolution of complex characters requires awareness of different degrees of morphostasis in character states as reflected in the unity and diversity of organisms. Evolutionary biologists who study groups of organisms with rich fossil records (e.g., molluscs, vertebrates, diatoms and foraminiferans) have the luxury of comparing both extinct and extant species to infer patterns of character evolution. Evolutionary biologists focused on microbial eukaryotes are most often restricted to comparisons of character states present in extant species to infer patterns of character evolution [1]. As expected, many species have retained traits that are completely compatible with those that are either inferred or observed to have been present in distant ancestors (e.g., Nautilus, Latimeria, Acanthostega, Diarthrognathus and Archaeopteryx; [2–4]). The discovery, characterization, and molecular phylogenetic analyses of species like these test hypotheses about trait evolution and illuminate the transitional stages that link divergent character states.
Among the most significant events in the evolutionary history of eukaryotes is the transition from a phagotrophic mode of life to a phototrophic mode of life, which has occurred several times independently [5–7]. This switch in nutritional mode has profound effects on the overall morphology and behavior of the descendant species. For instance, phagotrophic organisms tend to be highly motile in order to acquire food items and are usually composed of pliable cells or tissues that can accommodate ingested material; phototrophic organisms, by contrast, tend to be non-motile, sessile or planktonic and are usually composed of cells or tissues encased in a rigid wall for protection or structural integrity. The evolutionary transition from phagotrophy to phototrophy is therefore non-trivial, and the intermediate stages that bridge the morphological traits associated with these two lifestyles are important to understand. With this in mind, we have discovered and characterized a novel marine species with a suite of character states that completely conforms to the inferred transitional stage between the phagotrophic and phototrophic lifestyles present in a diverse group of microbial eukaryotes, namely the Euglenida.
A brief overview of euglenid biology and evolutionary history
The Euglenida is a large group of marine and freshwater flagellates that contains more than 1,000 described species with diverse modes of nutrition, including phagotrophy (bacterivory and eukaryovory), osmotrophy (sensu stricto) and phototrophy [8]. The best synapomorphy for this group of microbial eukaryotes is a cell surface consisting of longitudinally or helically arranged pellicle strips that articulate along their lateral margins [9]. Many species of phagotrophic, osmotrophic (sensu stricto), and phototrophic euglenids are capable of rhythmic cellular deformations called "euglenoid movement" (syn. "metaboly") that is facilitated by adjacent strips sliding past one another at their zones of articulation [8–13]. Most phagotrophic euglenids have a robust feeding apparatus comprised of two main bundles of microtubules called "rods" and four interior "vanes" that are organized like a pinwheel in transverse section [9, 14, 15]. The feeding apparatus is highly developed in phagotrophic euglenids and highly reduced in osmotrophic and phototrophic euglenids [16, 17].
Studies that have placed the morphological diversity of euglenids into a molecular phylogenetic context demonstrate that the mode of nutrition corresponds with general ultrastructural and behavioral traits [8, 9]. Although it is expected that euglenid cells are capable of absorbing nutrients from the environment regardless of other abilities to acquire nutrients (e.g., phagotrophy or phototrophy), euglenids can be separated into four functional groups based on distinct nutritional modes: bacterivores (cells that usually feed on bacteria-relatively small particles), eukaryovores (cells that usually feed on microbial eukaryotes-relatively large particles), primary osmotrophs (cells that have lost a feeding apparatus and are limited to the absorption of nutrients), and phototrophs (cells capable of acquiring nutrients through photosynthesis). Bacterivores and eukaryovores are each paraphyletic, and the latter evolved from the former; primary osmotrophs and phototrophs are monophyletic, and each group evolved from eukaryovorous ancestors independently [8, 9, 16, 18]. There are a couple of caveats to these designations: (1) the bacterivorous and eukaryovorous designations are not immune from exceptions (e.g., some Petalomonas species are relatively large and can consume yeast cells); (2) the loss of photosynthesis within phototrophic euglenids has led to several different lineages of "secondary" osmotrophs (cells that retain a non-photosynthetic plastid plus other ultrastructural features of phototrophic species; e.g., Euglena longa); and (3) the term "osmotrophy" refers to euglenid species that are limited to the absorption of nutrients and is not intended to imply that photosynthetic and phagotrophic euglenids are incapable of the absorbing nutrients from the environment.
Nonetheless, bacterivorous euglenids (e.g., Petalomonas, Ploeotia and Entosiphon) glide along substrates using one or two flagella, have a simple to complex feeding apparatus, and tend to be smaller with a rigid pellicle consisting of relatively few longitudinal strips (i.e., 4-12). Eukaryovorous euglenids (Peranema, Heteronema and Urceolus) also glide along substrates using one or two flagella, have a complex feeding apparatus, and tend to be larger with a flexible pellicle consisting of a relatively large number of helical strips (i.e., 16-60). The two main groups of euglenids that have lost phagotrophy, namely phototrophs and primary osmotrophs, contain lineages capable of euglenoid movement and several derived lineages that have lost euglenoid movement. The presence of euglenoid movement in some phototrophs and primary osmotrophs is inferred to reflect morphostasis from their eukaryovorous ancestors [8, 9, 13, 18]. Primary osmotrophic euglenids swim in the water column using one or two flagella, lack a feeding apparatus, and have either a flexible or a fused pellicle consisting of a relatively large number of helical strips (i.e., usually more than 16-30, Rhabdomonas costata has 6-8). Phototrophic euglenids are separated into two main groups: the Eutreptiales and the Euglenales. Members of the Eutreptiales are largely marine, swim in the water column using two or more flagella, contain chloroplasts, lack a conspicuous feeding apparatus, possess a pronounced photoreception apparatus, and have a flexible pellicle consisting of a relative large number of helical strips (i.e.,16-50). Members of the Euglenales are largely freshwater, normally swim in the water column using one emergent flagellum, contain chloroplasts, lack a conspicuous feeding apparatus, possess a pronounced photoreception apparatus, and have a flexible or rigid pellicle consisting of a relatively large number of (usually) helical strips (i.e., 16-120). The pellicle of phototrophic euglenids can be distinguished from all other euglenids by the presence of "posterior whorls of strip reduction" (i.e., patterns of strips that terminate before reaching the posterior end of the cell). The actual number of pellicle strips and posterior whorls of strip reduction present on any particular cell reflects phylogenetic relationships and fundamental developmental mechanisms in euglenids [8–10, 13, 19].
The chloroplasts of phototrophic euglenids originated through a secondary endosymbiotic relationship between eukaryovorous euglenids and green algae. The pellicle and feeding apparatus in species like Heteronema and Urceolus best approximate the inferred features present in the most recent non-photosynthetic ancestor of phototrophic euglenids [8, 13, 16]; the chloroplasts of prasinophyceans like Pyramimonas best approximate the inferred features present in the most recent ancestor of all euglenid chloroplasts [20–22]. Nonetheless, there are still substantial differences between the morphological and behavioral features of eukaryovorous euglenids and phototrophic euglenids that limit our ability to fully portray the evolutionary transition between these two very different lifestyles. An improved understanding of euglenid diversity is expected to help bridge remaining gaps in our knowledge of this key event. In this vein, we have discovered and comprehensively characterized a novel marine euglenid with a uniquely "mixotrophic" lifestyle; this species contains a functional chloroplast and habitually consumes a specific strain of green algae. In this context, the term "mixotrophy" refers to a euglenid that is capable of both phagotrophy and phototrophy. Behavioral and ultrastructural data derived from high resolution light microscopy, scanning and transmission electron microscopy (SEM and TEM), and molecular phylogenetic analysis of small subunit (SSU) rDNA sequences demonstrate that this novel species conforms to the expected transitional stage between phototrophic euglenids and eukaryovorous ancestors.
Methods
Collection of organism and maintenance of cultures
Seawater samples were collected from a tide pool at Pachena Beach, Bamfield, British Columbia, Canada (48° 47.551' N, 125° 06.974' W) on June 18, 2010. The samples were inoculated in Provasoli's Enriched Seawater (PES) medium [23] and maintained at room temperature for one week. Tetraselmis sp. was isolated from the enrichment culture by micropipetting into sterile PES medium. Rapaza viridis n. gen. et sp. was also isolated from the enrichment culture into PES medium containing Tetralselmis sp. as a food source. Both cultures were incubated at 20°C under an illumination of 55-59 μmol photons/m2/s with 12:12 light:dark (L:D) light regime. The cultures were transferred every 10 to 14 days by bringing 1 ml of the culture into 25 ml of PES medium and prey cells. The cultures of R. viridis n. gen. et sp. and Tetraselmis sp. have been deposited into the American Type Culture Collection (ATCC), Manasas, VA, USA as PRA-360 and PRA-361, respectively.
Light microscopy
Differential interference contrast (DIC) light micrographs were generated using a Zeiss Axioplan 2 imaging microscope equipped with Leica DC500 digital camera. Digital videos of live cells were generated using a Zeiss Axioplan 2 equipped with a Q imaging Microimager II digital camera and Q Capture v 2.8.1 software.
Growth experiments
Cells of R. viridis were exposed to several different food sources in addition to Tetraselmis sp. ATCC PRA-361. Cultures of Tetraselmis sp. NEPCC365, Tetraselmis sp. NEPCC498, T. striata NEPCC487 and T. tetrathelle NEPCC 483 were obtained from the Canadian Center for the Culture of Microorganisms (CCCM) at the University of British Columbia, Vancouver, BC, Canada. A culture of Navicula sp. ATCC PRA-314 was obtained from the American Type Culture Collection (Manasas VA, USA). Cultures of the prasinophyte Pycnococcus sp., the chlorophyte Dunaliella sp., and the dinoflagellate Scrippsiella trochoidea were established in the lab from different marine environments independently.
Cultures of R. viridis were starved of prey cells and observed every 2-3 days until the death of all R. viridis cells was confirmed. We confirmed the existence of at least one chloroplast in the starved R. viridis cells using a Zeiss Axioplan 2 imaging microscope after one week, three weeks and five weeks from the start of starvation.
Cultures of R. viridis were also grown in the dark at 20°C. Prey cells (Tetraselmis sp., ATCC PRA-361) were added every 1-2 days until the death of all R. viridis cells was confirmed. As a control experiment, cultures grown under normal light conditions were also treated exactly like the cultures grown in the dark (e.g., the same volume of prey cells was added at the same intervals of time).
Scanning electron microscopy
A culture of R. viridis was mixed in 1% (v/v) OsO4 in seawater at room temperature. The fixed cells were mounted on polycarbonate Millipore filters (13 mm diam., 5 μm pore size) or glass plates coated with poly-L-lysine at room temperature for 15 min. The cells were rinsed with distilled water and dehydrated with a graded ethanol series from 30% to absolute ethanol before being critical point dried with CO2 using a Tousimis Critical Point Dryer. The dried cells were then coated with gold using a Cressington 208HR High Resolution Sputter Coater and observed with a Hitachi S-4700 field emission SEM.
Transmission electron microscopy
Two different cultures were examined with transmission electron microscopy (TEM): (1) a culture starved of Tetraselmis sp. cells for three weeks and (2) a culture that was fed cells of Tetraselmis sp. one hour prior to fixation. The cultures were pre-fixed in 2.5% (v/v) glutaraldehyde with 0.2 M sucrose in 0.1 M sodium cacodylate buffer (SCB) (pH 7.2) at room temperature for 2 hours. The pre-fixed cells were washed in 0.2 M SCB (pH 7.2) twice and post-fixed in 1% (w/v) osmium tetroxide in 0.2 M SCB (pH 7.2) at room temperature for 1 hour. The fixed cells were dehydrated through a graded series of ethanol and 100% acetone. The dehydrated cells were then infiltrated with acetone-Epon 812 resin mixtures and 100% Epon 812 resin. Ultra-thin serial sections were collected on copper Formvar-coated slot grids, stained with 2% (w/v) uranyl acetate and lead citrate, and observed using a Hitachi H7600 TEM.
DNA extraction, PCR amplification, alignment and phylogenetic analysis
Genomic DNA was extracted using the MasterPure Complete DNA and RNA purification Kit (Epicentre, WI, USA) from a culture of R. viridis that was starved of prey cells for three weeks. Polymerase chain reactions (PCR) were performed using PuRe Taq Ready-To-Go PCR beads kit (GE Healthcare, Buckinghamshire, UK). The nearly complete eukaryotic SSU rDNA gene was amplified using the eukaryotic universal primers 5'-TGATCCTTCTGCAGGTTCACCTAC-3' and 5'-GCGCTACCTGGTTGATCCTGCCAGT-3' with the same PCR protocol described by Breglia et al. (2010) [24]. The amplified DNA fragments were purified from agarose gels using UltraClean 15 DNA Purification Kit (MO Bio, CA, USA), and subsequently cloned into the TOPO TA Cloning Kit (Invitrogen, CA, USA). One clone was sequenced using ABI Big-Dye reaction mix (BigDye 3.1) using the vector forward and reverse primers and also internal primers (nomet1134R: 5'-TTTAAGTTTCAGCCTTGCG-3' and SR4Eug: 5'-ACTGGAGGGCAAGYCTGGT-3') oriented in both directions. The new sequence was initially identified by BLAST analysis, confirmed with molecular phylogenetic analysis, and deposited into GenBank: AB679269.
The SSU rRNA sequence from R. viridis was added to a 39-taxon alignment focused on the diversity of euglenids using representative kinetoplastids and diplonemids as an outgroup. Ambiguously aligned positions and gaps were excluded from alignment, leaving 805 unambiguously aligned positions; the alignment is available from the authors upon request.
Maximum likelihood (ML) analysis was performed on the 39-taxon alignment using PAUP* version 4.0b10 [25]. Prior to starting the ML analysis, we used Akaike information criterion (AIC) test as implemented in the software jModelTest 0.1.1. [26] to find the model of evolution that best fits the data set for the Maximum likelihood (ML) analysis. The result indicated that the TIM1ef, which allows for equal base frequencies and four substitution rates (AC = GT; AT = CG; AG, CT) [27, 28], plus gamma model should be used for this data set. The parameters were as follows: assumed nucleotide frequencies are equal; substitution rate matrix with A-C substitutions = 1.0000, A-G = 2.7002, A-T = 0.7141, C-G = 0.7141, C-T = 3.8573, G-T = 1.0000; proportion of sites assumed to be invariable = 0 and rates for variable sites assumed to follow a gamma distribution with shape parameter = 0.3930. The ML tree was implemented using the heuristic search option with TBR branch swapping. Bootstrap analyses [29] were carried out for ML with 500 replicates to evaluate statistical reliability.
The alignment was also analyzed with Bayesian methods using the MrBayes 3.1.2 [30]. The program was set to operate the GTR model with a gamma distribution and four Monte-Carlo-Markov chains (MCMC) starting from a random tree. A total of 500,000 generations were calculated with trees sampled every 100 generations. The first 1,250 trees in each run were discarded as burn-in using the sumt command. Posterior probabilities correspond to the frequency at which a given node was found in the post burn-in trees.
Sequence availability
The SSU rDNA nucleotide sequences included in 39-taxon analyses for this paper are available from the GenBank database under the following accession numbers: Anisonema acinus [GenBank:AF403160], Bihospites bacati [GenBank:HM004354], Bodo saltans [GenBank:AY998648], Calkinsia aureus [GenBank:EU753419], Colacium sp. [GenBank:DQ140154], Dimastigella mimosa [GenBank:DQ207576], Dinema sulcatum [GenBank:AY061998], Diplonema ambulator [GenBank:AF380996], Diplonema papillatum [GenBank:AF119811], Discoplastis spathirhyncha [GenBank:AJ532454], Distigma proteus [GenBank:AF106036], Entosiphon sp. [GenBank:AY425008], Entosiphon sulcatum [GenBank:AF220826], Euglena gracilis [GenBank:AF283308], Euglena longa (as Astasia longa) [GenBank:AF112871], Euglena quartana (as Khawkinea quartana) [GenBank:U84732], Euglena stellata [GenBank:AF081590], Euglena viridis [GenBank:AF445460], Eutreptia viridis [GenBank:AF157312], Eutreptiella gymnastica [GenBank:AF081590], Eutreptiella pomquetensis [GenBank:AJ532398], Ichthyobodo necator [GenBank:AY224691], Lepocinclis buetschlii [GenBank:AF096993], Menoidium cultellus [GenBank:AF295019], Monomorphina sp. [GenBank:DQ140130], Neobodo designis [GenBank:AF209856], Notosoleus ostium [GenBank:AF403159], Peranema sp. [GenBank:AY048919], Peranema trichophorum [GenBank:AF386636], Petalomonas cantuscygni [GenBank:AF386635], Phacus aenigmaticus [GenBank:AF283313], Ploteotia costata [GenBank:AF525486], Rapaza viridis [GenBank: AB679269], Rhabdomonas costata [GenBank:AF295021], Rhynchomonas nasuta [GenBank:AY998642], Rhynchopus sp. [GenBank:AF380997], Strombomonas triquetra [GenBank:DQ140153], Trypanosoma evansi [GenBank:AY904050], Trypanosoma sp. [GenBank:EF375883].
Archiving
A digital archive of this paper is available from PubMed Central and print copies are available from libraries in the following five museums: Natural History Museum Library (Cromwell Road, London, SW7 5BD, UK), American Museum of Natural History (Department of Library Services, Central Park West at 79th St., New York, NY, 10024, USA), Muséum national d'Histoire naturelle (Direction des bibliothèques et de la documentation, 38 rue Geoffroy Saint-Hilaire, 75005 Paris, France), Russian Academy of Sciences (Library for Natural Sciences of the RAS Znamenka str., 11, Moscow, Russia) and Academia Sinica (Life Science Library, 128 Sec. 2 Academia Rd, Nankang Taipei 115, Taiwan R.O.C.).
Results
General morphology
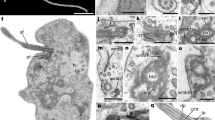
The relaxed, (well-fed) swimming cells of Rapaza viridis n. gen. et sp. were oval, 10.5-38.2 μm long (mean ± SD = 19.5 ± 7.3 μm, n = 50), and 2.9-15.1 μm wide (mean ± SD = 8.4 ± 3.3 μm, n = 50). The cells of R. viridis usually swam in a spiral pattern with the flagella moving rapidly and chaotically. Euglenoid movement was also observed when the cells were feeding on Tetraselmis or when the cells were pressured by a cover glass (Figure 1A, Additional file 1). Two flagella emerged from the flagellar pocket, were unequal in length, were adorned with hairs, and contained heteromorphic paraxial rods (Figures 1E, 2A and 3B). The longer anterior flagellum was about 1.25 times the length of the cell in the relaxed state and always directed forward. The shorter posterior flagellum was about 0.65 times the length of the relaxed cell and directed backward; this flagellum was sometimes directed forward and moved oar-like (Additional file 1). A robust anterior stigma that was independent of the chloroplasts was comprised of one to more than ten particles ranging from 0.8-1.3 μm in diam. (Figures 4B and 4D). A paraflagellar swelling formed a dense, lens-like body that was positioned within the flagellar pocket and against the stigma (Figure 4D). The cytoplasm contained several ellipsoid paramylon grains (Figure 1B), large Golgi bodies, and mitochondria with discoidal (paddle-shaped) cristae (Figure 4A). The typical euglenid nucleus contained permanently condensed chromosomes and a large central nucleolus (Figure 3A). The pellicle consisted of 16 helically arranged strips (Figures 4A and 5A) with delicate S-shaped frames that were supported by underlying microtubules (Figure 4C). Eight pellicle strips terminated before reaching the posterior end of the cell; these strips were organized in pairs that formed the four corners of a square-like pattern when the terminating strips were traced with lines (Figure 2D). The feeding apparatus consisted of a feeding pocket and one adjacent feeding rod comprised of 20 microtubules organized in four rows (4-6-6-4) (Figures 3B, C and 3D). We did not observe any vanes or amorphous material associated with these microtubules (Figure 3).
Differential interference contrast (DIC) light micrographs (LM) of Rapaza viridis n. gen. et sp. showing general morphological features of the species. A. Time series of a live cell showing euglenoid movement (arrow, stigma). B. LM showing paramylon grains (arrowheads) free in the cytoplasm. C. LM showing R. viridis capturing a Tetraselmis cell (arrowhead) with the anterior part of the cell. D. LM showing R. viridis engulfing a Tetraselmis cell. The arrow points to the theca of Tetraselmis, and the arrowheads denote the expanded cytostome of R. viridis. E-F. LMs of R. viridis cells that have been starved for one week (arrow, stigma; double arrowhead, euglenid chloroplast). G-H. LMs of R. viridis cells that were fed 24 hours earlier. E-H. same magnification. Scale bars 10 μm in A, C-H; 5 μm in B.
Scanning electron micrographs (SEM) of Rapaza viridis n. gen. et sp. A. SEM showing a relaxed elongated cell with helical pellicle strips and two flagella. The anterior flagellum (AF) and the posterior flagellum (PF) insert into the anterior part of the cell. B. SEM showing an R. viridis cell that has bored a hole into the cell surface of a captured Tetraselmis cell (arrowhead). The internal contents of the captured Tetraselmis cell (arrow) are being withdrawn by myzocytosis. C. SEM showing R. viridis engulfing a Tetraselmis cell (arrowhead). D. SEM of R. viridis showing posterior strip reduction (*), which connected by lines, forms a square-shaped whorl. Scale bars 5 μm in A-C; 2 μm in D.
Transmission electron micrographs (TEM) of Rapaza viridis n. gen. et sp. showing details of the feeding apparatus. A. Longitudinal TEM showing R. viridis capturing a Tetraselmis cell (arrow). The arrowhead indicates the theca (cell wall) of Tetraselmis, which is beginning to unravel. The chloroplast of R. viridis (Rcp) and the nucleus (N) containing permanently condensed chromosomes and a large nucleolus (Nu) is also visible. B. Transverse TEM through the flagellar pocket (flp) showing paraxial rods (PR), the pocket (fep) and the microtubules of the feeding rod (arrow). C. Transverse TEM through the feeding rod showing 20 microtubules organized in four rows (4-6-6-4). D. Longitudinal TEM through the feeding apparatus showing the feeding pocket ('fep' and arrows) and microtubules of the rod (arrowheads). Scale bars 5 μm in A; 1 μm in B and D; 200 nm in C.
Transmission electron micrographs (TEM) of Rapaza viridis n. gen. et sp. showing details of the cytoplasm, pellicle and photoreception apparatus. A. Transverse TEM showing the mitochondrion (M-arrowhead), the Golgi body (G-arrowhead) and 16 pellicle strips (arrows). B. Transverse TEM showing the stigma (arrows) and the paraflagellar swelling (Pa). C. Transverse TEM showing the S-shaped frames of the pellicle strips (arrowheads) and underlying microtubules (arrows). D. Sagittal TEM showing the stigma (arrows) and the paraflagellar swelling (Pa). Scale bars 1 μm in A, B and D; 500 nm in C.
Transmission electron micrographs (TEM) of Rapaza viridis n. gen. et sp. showing details of two types of chloroplasts. A. Transverse TEM showing the chloroplast of R. viridis and 16 pellicle strips (arrowheads). Pairs of thylakoid membranes (arrows) penetrate the pyrenoids (Py). The black arrow indicates the the pellicle strip that is shown in Figure 5B. B. Higher magnification TEM of Figure 5A focusing on the three membranes that surround the chloroplast of R. viridis (arrowheads) and the associated stacks of thylakoids (Ty). The black arrow indicates the the pellicle strip that is shown in Figure 5A. C. High magnification TEM showing electron-dense structures surrounded by a single membrane (arrowheads) that are situated near a chloroplast. D. TEM showing two different types of chloroplasts: The chloroplast of Tetraselmis sp. (Tcp) contains an eyespot (arrowheads); the chloroplast of R. viridis (Rcp) does not. E. Longitudinal TEM showing an engulfed Tetraselmis cell, including the nucleus (N), pyrenoid (Py) and associated starch (S) and chloroplast (Cp). Arrowheads indicate the pellicle strips of R. viridis. F. High magnification TEM showing the food vacuole of R. viridis (R-arrow) and the cell membrane of the engulfed Tetraselmis cell (T-arrow). White arrowheads mark the two membranes that surround the chloroplast of Tetraselmis sp. Black arrowheads mark the pellicle strips of R. viridis. G. High magnification TEM showing the eyespot (arrows) within the chlorplasts of Tetraselmis sp. Scale bars 1 μm in A, C and G; 200 nm in B and F; 4 μm in D and E.
Additional file 1: A movie showing euglenoid movement and flagellar motility in Rapaza viridis n. gen. et sp. (MOV 3 MB)
Feeding behavior
R. viridis ingested Tetraselmis cells when plentiful in the culture and were distinctly larger and bright green in the presence of food (Figures 1G and 1H). The Tetraselmis cells, including their chloroplasts, were completely digested over the course of about 12 hours. When starved of Tetraselmis prey, the cells of R. viridis became smaller and colorless, except for the retention of at least one intact chloroplast within the cytoplasm (Figures 1E and 1F). R. viridis could not survive for more than 35 days without exposure to a specific strain of Tetraselmis sp. (PRA-361). Different strains of Tetraselmis spp. and several other species of microalgae were added to the cultures in an attempt to grow R. viridis on an alternate food source: the prasinophytes Tetraselmis sp. NEPCC365, Tetraselmis sp. NEPCC498, T. striata NEPCC487, T. tetrathelle NEPCC 483, Pycnococcus sp., the diatom Navicula sp. ATCC PRA-314, the chlorophyte Dunaliella sp., and the dinoflagellate Scrippsiella trochoidea. R. viridis rejected all of these prey choices.
Adding Tetraselmis sp. (PRA-361) into a culture of starved R. viridis cells triggered a feeding frenzy. R. viridis captured Tetraselmis sp. (PRA-361) with the anterior part of the cell and either rotated rapidly or swam backward in a spiral pattern while dragging the prey cell (Figures 1C and 3A). Euglenoid movement was prominent during the processes of ingestion (Figures 1D, 2C and Additional file 2). R. viridis was capable of completely engulfing prey cells. In the process of engulfment, the theca (cell wall) of Tetraselmis was gradually loosened and peeled away by the repetitive peristaltic actions of euglenoid movement (Figures 1D and 2B-C). However, Tetraselmis cells with an intact theca could also be engulfed, but the theca was discharged from the anterior end of R. viridis cells soon afterwards. The continuous actions of euglenoid movement facilitated the complete ingestion and posterior transport of an engulfed Tetraselmis cell (Additional file 2). The entire process of engulfment took 5-40 minutes; at this point, the R. viridis cell would slow down euglenoid movement, elongate, and begin to swim again. Cells of R. viridis exposed to an ample food supply contained several ingested cells of Tetraselmis at one time.
Additional file 2: A movie showing Rapaza viridis n. gen. et sp. engulfing a cell of Tetraselmis sp. (1.7 MB MOV). This movie is three times faster than actual speed. (MOV 2 MB)
The chloroplasts of Rapaza viridis and Tetraselmis sp
There were two ultrastructurally distinct types of chloroplasts present in well-fed cells of R. viridis: (1) chloroplasts corresponding to the ultrastructure of green algae (i.e., Tetraselmis) and (2) chloroplasts corresponding to the ultrastructure of phototrophic euglenids. The euglenid-type chloroplasts were surrounded by three membranes, contained thylakoids in stacks of three, and contained one to three darkly stained pyrenoids without associated paramylon granules (Figures 5A, B and 5D). Double stacks of thylakoids also penetrated the pyrenoids in the euglenid-type chloroplasts (Figures 5A and 5D). Electron-dense structures surrounded by a single membrane were often observed just outside the euglenid chloroplast (Figure 5C). The Tetraselmis-type chloroplasts were often discernible within the cytoplasm of R. viridis by light microscopy because of their intraplastidial eyespots. The Tetraselmis-type chloroplasts were also distinguishable from the euglenid-type chloroplasts with TEM. A complete membrane surrounded engulfed Tetraselmis cells (Figure 5F). The pyrenoids in the Tetraselmis-type chloroplasts were surrounded by starch grains and did not contain penetrating thylakoids (Figure 5E). Two membranes, rather than three, surrounded the Tetraselmis-type chloroplasts (Figure 5F). Moreover, unlike the euglenid-type chloroplasts, intraplastidial eyespot globules were easily seen at the periphery of Tetraselmis-type chloroplasts (Figures 5D and 5G).
Starved cells of R. viridis almost always contained at least one intact chloroplast (Figures 1E and 1F). TEM demonstrated that this enduring chloroplast had the euglenid-type ultrastructure (Figure 5A). When cells of R. viridis were grown in the absence of light, they could not survive for more than one week. The result of this dark growth experiment was consistent even in the presence of an ample and continuous supply of Tetraselmis prey cells. Cells of R. viridis required both photosynthesis and (Tetraselmis) prey cells in order to survive.
Molecular phylogenetic position
We determined the nearly complete sequence of the SSU rRNA gene of R. viridis (2,669 bp). Molecular phylogenetic analysis of the 39-taxon alignment demonstrated that phototrophic euglenids (syn. euglenophytes), including secondary osmotrophic species (Euglena longa and E. quartana), formed a robust monophyletic group (100% bootstrap value and 1.00 Bayesian posterior probability) (Figure 6). R. virdis formed the nearest sister lineage to clade of phototrophic euglenids with relatively high statistical support (80% bootstrap value and 1.00 Bayesian posterior probability).
Phylogenetic position of Rapaza viridis n. gen. et sp. within the Euglenozoa as inferred from SSU rRNA gene sequences. Maximum-likelihood (ML) tree constructed using TIM1ef + G model of evolution on an alignment of 39 taxa and 805 unambiguously aligned sites, including 298 constant positions and 432 informative positions, using seven kinetoplastid and three diplonemid sequences as the outgroup. ML bootstrap values greater than 50% are shown. Thick branches indicate Bayesian posterior probabilities (PP) over 0.95. The branch leading to the fast-evolving Entosiphon clade has been shortened by one half (indicated by '1/2').
Discussion
Along with all of the expected euglenid features (e.g., pellicle strips, mitochondria with discoidal cristae, a nucleus with permanently condensed chromosomes, and paraxial rods), R. viridis possessed two heterodynamic flagella (rather than one emergent flagellum), swimming behavior, and euglenoid movement that was most reminiscent of members within the Eutreptiales. Like most species of Eutreptia and Eutreptiella, R. viridis lives in marine environments, which stands in contrast to the freshwater lifestyles of the vast majority of species within the Euglenales (e.g., Euglena, Phacus, Lepocinclis and Trachelomonas). However, R. viridis is clearly distinct from members of the Eutreptiales because of its eukaryovorous behavior and associated feeding apparatus. Moreover, the molecular phylogenetic analyses of SSU rDNA sequences supports the placement of R. viridis as the nearest sister lineage to all photosynthetic euglenids (i.e., the clade consisting of the Eutreptiales and the Euglenales) rather than sister to or within the Eutreptiales. R. viridis is also clearly distinct from other eukaryovorous euglenids in having features present in all other phototrophic euglenids, such as functional chloroplasts and a robust plastid-independent photoreception apparatus (i.e., a stigma plus a photosensory swelling at the base of the dorsal flagellum). Although there is inconclusive evidence that the eukaryovore Urceolus cyclostomus might also possess a weakly developed photoreception apparatus [16], R. viridis represents the first indisputable example of a eukaryovorous euglenid with photosensory ability. This would enable R. viridis to maintain its position in the water column in order to increase the likelihood of encountering its preferred photosynthetic prey cells (Tetraselmis sp.). However, because R. viridis is also photosynthetic, the photosensory ability could have more to do with maintaining an optimal position in the light for its own photosynthesis. Overall, the molecular phylogenetic position and combination of morphological and behavioral traits in R. viridis is more than just novel; these data completely conform to the inferred ancestral traits that existed during the evolutionary transition from eukaryovorous lifestyles to phototrophic lifestyles.
Transitional character states
The molecular phylogenetic analysis of SSU rDNA placed R. viridis squarely between phagotrophic and phototrophic euglenids as the nearest sister lineage to the entire euglenophyte clade (i.e., all phototrophic euglenids and their secondary osmotrophic descendants). This phylogenetic context alone makes R. viridis a particularly intriguing candidate for gaining new insights into euglenid character evolution, especially in regard to the endosymbiotic origin of chloroplasts. The ultrastructural and behavioral features in R. viridis were perhaps even more compelling because they were so intermediary between the features already described in phagotrophic and phototrophic euglenids (Figure 7); this is especially relevant for characters associated with modes of nutrition, the feeding apparatus, the flagellar apparatus and the pellicle.
Illustrations showing the transitional character states present in Rapaza viridis n. gen. et sp. that link phagotrophic euglenids with phototrophic euglenids. Illustrations of five different euglend cells emphasizing the size of the feeding apparatus, the number of flagella, and the presence or absence of chloroplasts, pyrenoids, paramylon grains and a stigma. The illustrations below each representative cell show the number of pellicle strips and whorls of posterior strip reduction as indicated by different colors. Pellicle strips that reach the posterior end of the cell are shown in yellow; strips that terminate before reaching the posterior end are shown in blue and purple. The solid grey lines that connect the terminating strips represent each exponential whorl of reduction. Note that the cell representing the Eutreptiales has one zigzag (exponential) whorl associated with two staggered subwhorls. The illustrations are based on the ultrastructural studies of Ploeotia [45–47], Heteronema scaphurum [48], the Eutreptiales (e.g., Eutreptia pertyi) [14, 19, 36, 49], and the Euglenales (e.g., Euglena laciniata for the pattern of pellicle strips and Eugelna gracilis for general shield-shaped plastids) [19, 50, 51].
Mixotrophy
R. viridis was unable to survive when placed in the dark or when deprived of Tetraselmis prey cells; therefore, R. viridis required both photosynthesis and phagotrophy to satisfy its overall nutritional requirements. This "mixotrophic" mode of nutrition is rare and has only been described in a few groups of eukaryotes, such as in some dinoflagellates, haptophytes and stramenopiles [31–33]. Until now, mixotrophy was completely unknown within the Euglenida. Nonetheless, a mixotrophic mode of nutrition is a logical transitional stage during the establishment of endosymbiotic partnerships between (ancestrally) phagotrophic host cells and phototrophic prey cells (Figure 7). Once the required horizontal gene transfer events and gene retargeting mechanisms established a permanent photosynthetic organelle within a new host cell, the ability to lose phagotrophy and to rely exclusively on phototrophy becomes possible [34, 35]. This has clearly happened in several different lineages of eukaryotes independently, including in the most recent ancestor of all phototrophic euglenids [8]. Therefore, the mixotrophic mode of nutrition in R. viridis is entirely consistent with the molecular phylogenetic position of this species and clarifies an important transitional stage in the origin of phototrophy in euglenids (Figure 7).
Feeding apparatus
The main components of the feeding apparatus in most phagotrophic euglenids are two robust rods that can extend the entire length of the cell. Phototrophic euglenids have retained a highly reduced feeding apparatus without rods [9, 17]. A few phototrophic euglenids within the Eutreptiales, however, have retained small clusters of microtubules that are inferred to be homologous to the rods of phagotrophic euglenids [36, 37]. The feeding rod we characterized in R. viridis is intermediate in size and organization between the robust rods in phagotrophic species and the reduced rods in the Eutreptiales (e.g., Eutreptiella eupharyngea) (Figure 7). This result is entirely consistent with the molecular phylogenetic position of this species, substantiates the above homology statement, and clarifies an important transitional stage in the origin of phototrophy in euglenids (Figure 7).
Flagellar apparatus
Phagotrophic euglenids generally glide along surfaces with two relatively thick heterodynamic flagella: the anterior (dorsal) flagellum is directed forward and twitches at the tip; the posterior (ventral) flagellum is directed backwards and glides against the substrate [8]. Phototrophic (and osmotrophic) euglenids generally swim in the water column using one or two flagella that move somewhat erratically or move in a controlled figure-eight configuration that pulls the cell forward. These modes of swimming allow phototrophic euglenids to exploit the water column above the substrate [9]. The cells of R. viridis were capable of swimming in the water column in a pattern that was most reminiscent of species within the Eutreptiales. Therefore, R. viridis represents the first example of a eukaryovorous euglenid that is also capable of swimming in the water column; this is another feature that is consistent with the molecular phylogenetic position of this species and that clarifies an important transitional stage in the origin of phototrophy in euglenids (Figure 7).
Pellicle
The substructure, number, and organization of pellicle strips reflects phylogenetic relationships within euglenids [8–11, 13, 19]. For instance, members of the Euglenales tend to have strips with relatively thick proteinaceous frames and underlying projections that interconnect adjacent strips [9, 11, 13, 38]. Phagotrophic species, (plastic) primary osmotrophic species, and members of the Eutreptiales have relatively thin proteinaceous frames without projections. The strip substructure of R. viridis was most similar to the strips in members of the Eutreptiales and eukaryovores like Dinema and Peranema, which is consistent with the transitional state expected between eukaryovorous and phototrophic euglenids [11, 13]. The total number of strips in R. viridis (16) was relatively low in comparison to Peranema and Eutreptia and more consistent with the number of strips found in eukaryovores like Dinema and primary osmotrophs like Distigma (16-22) [10, 16].
Posterior whorls of strip reduction have only been observed in phototrophic euglenids; however, some relatively obscure patterns of posterior strip reduction have also been observed in one eukaryovore (Peranema; [13]). The number and pattern of posterior whorls of strip reduction varies in different species and reflects phylogenetic relationships within phototrophic euglenids [19, 39]. Posterior whorls of strip reduction are usually "exponential" (i.e., the number of strips that terminate on a whorl equals the number of strips that pass through the whorl, leading to a pattern whereby the number of strips is halved at each whorl), but in some species the whorls are separated into two or more "linear" subwhorls (i.e., the number of strips that terminate is the same for all subwhorls; Figure 7) [10, 13, 39]. The range for the number of exponential whorls of strip reduction known so far in the Euglenales is two to four [19]. One member of the Eutreptiales (Eutreptia pertyi) has been shown to possess two linear subwhorls of posterior strip reduction that corresponds to one exponential whorl of strip reduction [13] (Figure 7). Aside from a few derived species of Phacus [40], R. viridis is the first and only euglenid described so far with one clear exponential whorl of strip reduction; this precisely corresponds to the expected character state that was present in the most recent ancestor of all euglenids possessing posterior whorls of strip reduction (i.e., all phototrophic species) [9, 13, 19]. The eight strips that terminate on the whorl of strip reduction in R. viridis form four distinct pairs that, when connected by lines, define the equidistant corners of a square (Figure 2D). This pattern of posterior strip reduction is novel among all euglenids characterized so far.
The chloroplasts of Rapaza viridis
Some dinoflagellates, animals, and foraminiferans possess "kleptochloroplasts", which are transiently functional chloroplasts derived from algal food. The host cells retains these chloroplasts for a short period of time in order to obtain products of photosynthesis as a source of nutrition; hosts are not capable of maintaining chloroplasts as permanent organelles that can be inherited from generation to generation. Kleptochloroplasts are ultimately digested, and the host organism needs to replenish them by regularly consuming algal food. Eukaryovorous euglenids (e.g., Peranema and Urceolus) do not have chloroplasts but they often engulf microalgae as their primary source of food, so well-fed cells can superficially appear full of chloroplasts; these chloroplasts disappear when the eukaryovorous cell is starved. By contrast, the cells of R. viridis always retained at least one chloroplast even when starved for over a month and cannot survive in the dark even when regularly exposed to an abundant food supply. Therefore, cells of R. viridis require both photosynthesis and (Tetraselmis) prey cells in order to survive.
Our ultrastructural observations of well-fed and starved cells of R. viridis demonstrated that this species usually contains both stable chloroplasts and transient Tetraselmis chloroplasts that can be readily distinguished from one another. The stable chloroplasts in R. viridis were surrounded by three membranes, lacked an intraplastidial eyespot and contained pyrenoids penetrated by thylakoid membranes (Figures 5A, B and 5D). These features are consistent with the ultrastructure of the chloroplasts in many species of phototrophic euglenids, such as Colacium cyclopicolum and Euglena deses [41, 42]. However, the presence of more than one pyrenoid per chloroplast in some cells was a novel feature of R. viridis that has not been observed in any other species of euglenophyte so far (Figures 3A and 5A).
Nonetheless, euglenid chloroplasts arose as a consequence of an endosymbiotic relationship between a eukaryovorous euglenid and green algal prey cells that were most similar to prasinophyceans [9, 20–22]. The fact that our isolate requires both photosynthesis and (Tetraselmis) prey cells in order to survive provides additional support for this inference. However, the relatively close relationship between the stable, euglenid-type chloroplasts and the transient Tetraselmis chloroplasts in R. viridis might create additional challenges when characterizing each of them at the genomic level. Nonetheless, the molecular phylogenetic position of R. viridis suggests that the genome of their stable, euglenid-type chloroplasts will offer some of the most compelling insights into the earliest evolutionary stages in the endosymbiotic acquisition of chloroplasts in euglenids and beyond.
Conclusion
The unusual combination of features in R. viridis combined with its molecular phylogenetic position completely conforms to the expected transitional stage that occurred during the early evolution of phototrophic euglenids from phagotrophic ancestors. The marine mixotrophic mode of nutrition, the preference for green algal prey cells, the structure of the feeding apparatus, and the organization of the pellicle are outstanding examples of morphostasis that clarify pivotal stages in the evolutionary history of this diverse group of microbial eukaryotes.
Formal taxonomic descriptions
Euglenozoa [43]
Euglenida [44] (ICZN)
Rapaza n. gen. Yamaguchi, Yubuki & Leander 2012
Description
Cells solitary and mixotrophic. Two heterodynamic flagella, unequal in length. Euglenoid movement with helically arranged pellicle strips. A minimum of one discoidal chloroplast surrounded by three membranes and with pyrenoids penetrated by double stacks of thylakoids. Robust extra-plastidic stigma and paraflagellar swelling. Eukaryovorous on microalgae using a feeding apparatus consisting of one rod of microtubules, a feeding pocket, and no vanes.
Type species
Rapaza viridis Yamaguchi, Yubuki & Leander 2012
Etymology
Latin "rapax", meaning seizing and grasping in reference to the feeding behavior of the cell. The adjective 'rapax' is used as substantive noun. Feminine.
Rapaza viridis n. sp. Yamaguchi, Yubuki & Leander 2012
Description
Swimming cells are slender with a tapered posterior end, 10.5-38.2 μm long (average, 19.5 μm), 2.9-15.1 μm wide (average, 8.4 μm); with two heterodynamic flagella, unequal in length, same in thickness; the longer flagellum about twice the length of the shorter flagellum; with stigma comprised of several (1 to more than 10) pigmented particles at the anterior part of the cell; ellipsoid paramylon grains free in the cytoplasm; pellicle consisting of 16 helical strips; one square-shaped whorl of exponential strip reduction; feeding apparatus comprised of a feeding rod consisting of four rows of 20 microtubules (4-6-6-4); preferred prey Tetraselmis sp.
Holotype and hapantotypes
Both resin-embedded cells used for TEM and cells on gold sputter-coated SEM stubs have been deposited in the Beaty Biodiversity Research Centre (Marine Invertebrate Collection; accession number MI-PR113) at the University of British Columbia, Vancouver, Canada. All figures in the manuscript are based on the authentic culture, and Figure 1A has been selected as the holotype.
DNA sequence
A sequence of the small subunit rRNA gene is deposited as GenBank Accession No. AB679269).
Type locality
Pachena Beach, Bamfield, British Columbia, Canada (48° 47.551' N, 125° 06.974' W), June 18, 2010.
Habitat
Tide pools, marine.
Authentic culture
PRA-360. This culture is maintained in the American Type Culture Collection (ATCC), Manasas VA, USA.
Etymology
The specific epithet, viridis (green), refers to the color of the stable chloroplasts and prey cells. The binomial is Latin for "green grasper".
Abbreviations
- AF:
-
anterior flagellum
- fep:
-
feeding pocket
- flp:
-
flagellar pocket
- G:
-
Golgi body
- LM:
-
light microscope
- M:
-
mitochondrion
- N:
-
nucleus
- Nu:
-
nucleolus
- Pa:
-
paraflagellar swelling
- PES:
-
Provasoli's Enriched Seawater
- PF:
-
posterior flagellum
- PR:
-
paraxial rod
- Rcp:
-
chloroplast of R. viridis
- Py:
-
pyrenoid
- S:
-
starch grain
- SCB:
-
sodium cacodylate buffer
- SEM:
-
scanning electron microscope
- Tcp:
-
chloroplast of Tetraselmis sp.
- TEM:
-
transmission electron microscope
- Ty:
-
thylakoids in stacks.
References
Leander BS, Keeling PJ: Morphostasis in alveolate evolution. Trends Ecol Evol. 2003, 18: 395-402. 10.1016/S0169-5347(03)00152-6.
Johanson Z, Long JA, Talent JA, Janvier P, Warren JW: Oldest coelacanth, from the Early Devonian of Australia. Biol Lett. 2006, 2: 443-446. 10.1098/rsbl.2006.0470.
Clack JA: The fin to limb transition: new data, interpretations, and hypotheses from paleontology and developmental biology. Annu Rev Earth Planet Sci. 2009, 37: 163-179. 10.1146/annurev.earth.36.031207.124146.
Xu X, You H, Du K, Han F: An Archaeopteryx like theropod from China and the origin of Avialae. Nature. 2011, 475: 465-470. 10.1038/nature10288.
Reyes-Prieto A, Weber AP, Bhattacharya D: The Origin and Establishment of the Plastid in Algae and Plants. Annu Rev Genet. 2007, 41: 147-168. 10.1146/annurev.genet.41.110306.130134.
Lane CE, Archibald JM: The eukaryotic Tree of Life: endosymbiosis takes its TOL. Trends Ecol Evol. 2008, 23: 268-275. 10.1016/j.tree.2008.02.004.
Keeling PJ: The endosymbiotic origin, diversification and fate of plastids. Phil Trans R Soc B. 2010, 365: 729-748. 10.1098/rstb.2009.0103.
Leander BS: Did trypanosomatid parasites have Phototrophic ancestors?. Trends Microbiol. 2004, 12: 251-258. 10.1016/j.tim.2004.04.001.
Leander BS, Esson HJ, Breglia SA: Macroevolution of complex cytoskeletal systems in euglenids. Bioessays. 2007, 29: 987-1000. 10.1002/bies.20645.
Leander BS, Farmer MA: Comparative morphology of the euglenid pellicle. I. Patterns of strips and pores. J Eukaryot Microbiol. 2000, 47: 469-10.1111/j.1550-7408.2000.tb00076.x.
Leander BS, Farmer MA: Comparative morphology of the euglenid pellicle. II. Diversity of strip substructure. J Eukaryot Microbiol. 2001, 48: 202-217. 10.1111/j.1550-7408.2001.tb00304.x.
Leander BS, Farmer MA: Evolution of Phacu (Euglenophyceae) as inferred from pellicle morphology and small subunit rDNA. J Phycol. 2001, 37: 143-159. 10.1046/j.1529-8817.2001.037001143.x.
Leander BS, Witek RP, Farmer MA: Trends in the evolution of the euglenid pellicle. Evolution. 2001, 55: 2115-2135.
Triemer RE, Lawrence F: Structure and operation of the feeding apparatus in a colorless euglenoid, Entosiphon sulcatu. J Protozoology. 1987, 34: 39-47.
Triemer RE, Farmer MA: An ultrastructural comparison of the mitotic apparatus, feeding apparatus, flagellar apparatus and cytoskeleton in euglenids and kinetoplastids. Protoplasma. 1991, 164: 91-104. 10.1007/BF01320817.
Leander BS, Triemer RE, Farmer MA: Character evolution in heterotrophic euglenids. Eur J Protistol. 2001, 37: 337-356. 10.1078/0932-4739-00842.
Shin W, Brosnan S, Triemer RE: Are cytoplasmic pockets (MTR/pocket) present in al photosynthetic euglenoid genera?. J Phycol. 2002, 38: 790-799. 10.1046/j.1529-8817.2002.01208.x.
Busse I, Preisfeld A: Systematics of primary osmotrophic euglenids: A molecular approach to the phylogeny of Distigm and Astasi. Int J Syst Evol Microbiol. 2002, 53: 617-624.
Esson HJ, Leander BS: A model for the morphogenesis of strip reduction patterns in phototrophic euglenids: Evidence for heterochrony in pellicle evolution. Evol Dev. 2006, 8: 378-388. 10.1111/j.1525-142X.2006.00110.x.
Ishida K, Cao Y, Hasegawa M, Okada N, Hara Y: The origin of chlorarachniophyte plastids, as inferred from phylogenetic comparisons of amino acid sequences of EF-Tu. J Mol Evol. 1997, 45: 682-687. 10.1007/PL00006272.
Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ: The complete chloroplast genome of the chlorarachniophyte Bigelowiella natan: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007, 24: 54-62.
Turmel M, Gagnon MC, O'Kelly CJ, Otis C, Lemieux C: The chloroplast genomes of the green algae Pyramimona, Monomasti, and Pycnococcu shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 2009, 26: 631-648.
Provasoli L: Media and prospects for the cultivation of marine algae. Cultures and Collections of Algae. Proceedings of the U.S.-Japan Conference. Japanese Society for Plant Physiology: September 1966; Hakone. Edited by: Watanabe A, Hattori A. 1968, 63-75.
Breglia SA, Yubuki N, Hoppenrath M, Leander BS: Ultrastructure and molecular phylogenetic position of a novel euglenozoan with extrusive episymbiotic bacteria: Bihospites bacat n. gen. et sp. (Symbiontida). BMC Microbiol. 2010, 10: 145-10.1186/1471-2180-10-145.
Swofford DL: PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. 2002, Sunderland, MA: Sinauer
Posada D: jModelTest: Phylogenetic Model Averaging. Mol Biol Evol. 2008, 25: 1253-1256. 10.1093/molbev/msn083.
Posada D: Using Modeltest and PAUP* to select a model of nucleotide substitution. Current Protocols in Bioinformatics. Edited by: Baxevanis AD, Davison DB, Page RDM, Petsko GA, Stein LD, Stormo GD. 2003, New York: John Wiley & Sons, Inc, 6.5.1-6.5.14.
Posada D: Selection of models of DNA evolution with jModelTest. Bioinformatic for DNA sequence analysis. Edited by: Posada D. 2009, New York: Humana Press, 93-112.
Felsenstein J: Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985, 39: 783-791. 10.2307/2408678.
Huelsenbeck JP, Ronquist F: MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755. 10.1093/bioinformatics/17.8.754.
Holen DA, Boraas ME: Mixotrphy in chrysophtes. Chrysophyte Algae: Ecology, Phylogeny and Development. Edited by: Sandgren CD, Smol JP, Kristiansen J. 1995, Cambridge: Cambridge University Press, 119-140.
Stoecker DK, Tillmann U, Granéli E: Phagotrophy in harmful algae. Ecology of Harmful Algae. Edited by: Granéli E, Turner JT. 2006, Berlin: Springer-Verlag, 177-187.
Hansen PJ: The role of photosynthesis and food uptake for the growth of marine mixotrophic dinoflagellates. J Eukaryot Microbiol. 2011, 58: 203-214. 10.1111/j.1550-7408.2011.00537.x.
Archibald JM, Rogers M, Toop M, Ishida K, Keeling PJ: Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natan. Proc Natl Acad Sci USA. 2003, 100: 7678-7683. 10.1073/pnas.1230951100.
Keeling PJ, Archibald JM: Organelle Evolution: what's in a name?. Curr Biol. 2008, 18: R345-R347. 10.1016/j.cub.2008.02.065.
Walne PL, Moestrup Ø, Norris RE, Ettl H: Light and electron microscopical studies of Eutreptiella eupharynge sp. nov. (Euglenophyceae) from Danish and American waters. Phycologia. 1986, 25: 109-126. 10.2216/i0031-8884-25-1-109.1.
Triemer RE, Lewandowski CL: Ultrastructure of the basal apparatus and putative vestigial feeding apparatuses in a quadriflagellate euglenoid (Euglenophyta). J Phycol. 1994, 30: 28-38. 10.1111/j.0022-3646.1994.00028.x.
Esson HJ, Leander BS: Visualizing the complex substructure of euglenid pellicle strips with SEM. Phycologia. 2008, 47: 529-532. 10.2216/08-26.1.
Esson HJ, Leander BS: Novel pellicle surface patterns on Euglena obtus Schmitz (Euglenophyta), a euglenophyte from a benthic marine environment: Implications for pellicle development and evolution. J Phycol. 2008, 44: 132-141. 10.1111/j.1529-8817.2007.00447.x.
Esson HJ, Leander BS: Evolution of distorted pellicle patterns in rigid photosynthetic euglenids (Phacu Dujardin). J Eukaryot Microbiol. 2010, 57: 19-32. 10.1111/j.1550-7408.2009.00447.x.
Leedale GF: Euglenoid Flagellates. 1967, New Jersey: Prentice-Hall Inc
Zakrys B, Cambra-Sanchez J, Walne PL: Chloroplast ultrastructure of Euglena cuneat Pringsheim, E. dese Ehrenberg and E. mutabili (Euglenophyceae): Taxonomic Significance. Acta Protozool. 2001, 40: 161-167.
Cavalier-Smith T: Eukaryote kingdoms: seven or nine?. Biosystems. 1981, 14: 461-481. 10.1016/0303-2647(81)90050-2.
Bütschli O: Mastigophora. Dr H.G. Bronn's Klassen und Ordnungen des Thier-Reichs, Volume 1. Protozoa: No. 2. 1884, Leipzig: C.F. Winter, 617-1097.
Triemer RE, Farmer MA: The ultrastructural organization of the heterotrophic euglenids and its evolutionary implications. The Biology of Free-living Heterotrophic Flagellates. Edited by: Patterson DJ, Larsen J. 1991, Oxford: Clarendon Press, 185-204.
Belhadri A, Bayle D, Brugerolle G: Biochemical and immunological characterization of intermicrotubular cement in the feeding apparatus of phagotrophic euglenoids: Entosipho, Peranem, and Ploeoti. Protoplasma. 1992, 168: 113-124. 10.1007/BF01666257.
Linton EW, Triemer RE: Reconstruction of the flagellar apparatus in Ploeotia costat (Euglenozoa) and its relationship to other euglenoid flagellar apparatuses. J Eukaryot Microbiol. 2001, 48: 88-94. 10.1111/j.1550-7408.2001.tb00419.x.
Breglia SA, Yubuki N, Leander BS: Ultrastructure and molecular phylogenetic position of Heteronema scaphurum: A eukaryovorous euglenid with a cytoproct. J Eukaryot Microbiol. 2012,
Solomon JA, Walne PL, Dawson NS, Willey RL: Structural characterization of Eutrepti (Euglenophyta). II. The flagellar root system and putative vestigial cytopharynx. Phycologia. 1991, 30: 402-10.2216/i0031-8884-30-5-402.1.
Surek B, Melkonian M: A cryptic cytostome is present in Euglen. Protoplasma. 1986, 133: 39-49. 10.1007/BF01293186.
Shin W, Boo SM, Inouye I: Ultrastructure of Euglena anabaen var. mino (Euglenophyceae). Phycol Res. 2000, 48: 19-25. 10.1111/j.1440-1835.2000.tb00126.x.
Acknowledgements
This research was supported by grants from the Tula Foundation (Centre for Microbial Diversity and Evolution), the National Science and Engineering Research Council of Canada (NSERC 283091-09), the National Science Foundation (Assembling the Tree of Life NSF #EF-0629624), and the Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity. We also thank Erick James for access and assistance with the digital movies. We also thank the UBC Bioimaging Facility for technical assistance with the SEM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AY and NY collected the water samples from Pachena Beach, British Columbia; established and maintained cultures of Rapaza virids and Tetraselmis sp.; generated the LM, SEM, TEM and SSU rDNA sequence data; and drafted an initial version of the manuscript. BSL funded, facilitated, and supervised the collection, interpretation and presentation of the behavioral, ultrastructural, and molecular phylogenetic data and wrote subsequent versions of the manuscript. All authors have read, edited and approved the final manuscript.
Aika Yamaguchi, Naoji Yubuki and Brian S Leander contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yamaguchi, A., Yubuki, N. & Leander, B.S. Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: description of Rapaza viridis n. gen. et sp. (Euglenozoa, Euglenida). BMC Evol Biol 12, 29 (2012). https://doi.org/10.1186/1471-2148-12-29
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-12-29