Abstract
Background
The Galliformes is a well-known and widely distributed Order in Aves. The phylogenetic relationships of galliform birds, especially the turkeys, grouse, chickens, quails, and pheasants, have been studied intensively, likely because of their close association with humans. Despite extensive studies, convergent morphological evolution and rapid radiation have resulted in conflicting hypotheses of phylogenetic relationships. Many internal nodes have remained ambiguous.
Results
We analyzed the complete mitochondrial (mt) genomes from 34 galliform species, including 14 new mt genomes and 20 published mt genomes, and obtained a single, robust tree. Most of the internal branches were relatively short and the terminal branches long suggesting an ancient, rapid radiation. The Megapodiidae formed the sister group to all other galliforms, followed in sequence by the Cracidae, Odontophoridae and Numididae. The remaining clade included the Phasianidae, Tetraonidae and Meleagrididae. The genus Arborophila was the sister group of the remaining taxa followed by Polyplectron. This was followed by two major clades: ((((Gallus, Bambusicola) Francolinus) (Coturnix, Alectoris)) Pavo) and (((((((Chrysolophus, Phasianus) Lophura) Syrmaticus) Perdix) Pucrasia) (Meleagris, Bonasa)) ((Lophophorus, Tetraophasis) Tragopan))).
Conclusions
The traditional hypothesis of monophyletic lineages of pheasants, partridges, peafowls and tragopans was not supported in this study. Mitogenomic analyses recovered robust phylogenetic relationships and suggested that the Galliformes formed a model group for the study of morphological and behavioral evolution.
Similar content being viewed by others
Background
The Galliformes, a well-known and widely distributed Order in Aves, contains about 70 genera and more than 250 species including the domestic chicken (Gallus gallus), green peacock (Pavo muticus) and turkey (Meleagris gallopavo), among others. Many galliforms have beautiful ornamentations and they play an important role in hunting and entertainment. Regardless, these birds are best known for their importance in agriculture and as model organisms in scientific studies [1–6].
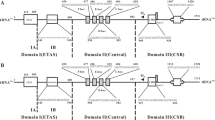
The phylogenetic relationships of galliforms have long been the focus of research [7–21]. Traditionally, the Galliformes contained seven families: Megapodiidae (scrubfowl and brush-turkeys), Cracidae (curassows and guans), Tetraonidae (grouse), Meleagrididae (turkey), Numididae (guineafowls), Odontophoridae (New World quails) and Phasianidae (pheasants and Old World quails). Phasianids formed the most diverse and complex group, including two lineages: the pheasants and Old World partridges (OW quails). The pheasants were further divided into four lineages: peafowls, gallopheasants, junglefowls and tragopans [22] (Fig. 1a). However, this taxonomy was not supported by recent molecular and morphological analyses (Fig. 1b, c, d), which failed to obtain a consistent result and added more fuel to an already heated debate.
Phylogenetic hypotheses from various molecular, morphological and behavioral analyses of gamebirds. (a) The traditional classification from Johnsgard (1986); (b) morphological and behavioral data (Dyke et al., 2003); (c) combined data including two mitochondrial genes (CytB, ND2) and four nuclear introns (BFib7, DCoH3, OvoG, and Rhod1) (Kimball and Braun 2008); (d) insertion events of CR1 retrotransposable elements (Kaiser et al., 2007).
Mitochondrial DNA analyses revealed that Gallus (Tribe Phasianini), Francolinus (Tribe Perdicini) and Bambusicola (Tribe Perdicni) clustered together, suggesting that pheasants and partridges were not monophyletic groups [9, 12]. Subsequently, these findings were supported by the analyses of nuclear gene sequences [11], combined mt genes and nuclear gene sequences (Fig. 1c) [17, 20] and sequences from retrotransposable elements (Fig. 1d) [19]. However, one morphological study [15] conflicted with these assessments (Fig. 1b). The phylogenetic relationships within the Galliformes, and especially the unsolved branching order in the Phasianidae, limited interpretations of their morphological and ecological convergent evolution. The absence of a phylogeny also impacted on conservation initiatives.
Internal nodes resolved by previous studies were generally very short. Further, bootstrap support values were very low and many species had unresolved relationships, suggesting a rapid radiation of the Phasianidae [8, 9]. Although the resolution of branching orders during a rapid radiation has proven to be challenging, large DNA sequence datasets have a much higher probability of recovering a robust tree [23–27].
Recently, mt genomes have been widely used to reconstruct intractable phylogenies [28–33]. In general, mtDNA accumulates mutations at a relatively faster rate than nuclear DNA, thus making it particularly useful for revealing closely spaced branching events. Considering that previous phylogenetic studies based on a single gene or a few genes failed to resolve the internal branching orders within the Galliformes, we sequenced the complete mt genomes of 14 galliform birds, and obtained other mt genomes from GenBank (Additional file 1). Consequently, we used extensive mt genomes from major groups of galliform birds to infer their phylogenetic relationships.
Results
Characteristics of the mitochondrial genome
The general characteristics of mt genomes of 34 galliforms and six anserforms are summarized in Additional file 1. The lengths of the complete mt genomes range from 16,604 to 16,870 bp. Length differences are mainly due to variation in the Control Region (CR). The overall average nucleotide composition was A = 29.3%, C = 30.65%, G = 14.51% and T = 25.45%. All mitochondrial gene organizations conformed to the standard avian gene order (chicken) [31, 34].
Phylogenetic relationships
Tests for stationarity of base composition among the taxa for each mt gene revealed that the Galliformes did not differ significantly in their base content (P > 0.05). Only Alectoris chukar for CoxI and ND5, Alectoris lathami for ND5, and Bonasa bonasia for CoxII, ND2 and ND4 differed marginally significantly. However, all five outgroup taxa failed the stationarity tests for some data partitions as follows: Anas platyrhynchos for CoxI, CR, ND2 and ND5; Anser albifrons for CR, ND2 and ND5; Anseranas semipalmata for CR; Aythya americana for CoxI, CR, ND2 and ND5; Branta canadensis for CoxI, CR and ND5; and Cygnus columbianus for CR (Additional file 2). In summary, CR had the greatest number of cases (in all five outgroups) that deviated from stationarity, and Anas platyrhynchos and Aythya americana deviated from stationarity most frequently (four times).
Because we mainly used the combined sequences to reconstruct the tree, we also calculated base composition for the 12 protein-coding gene set (Additional file 3). We found that base composition was not significantly different in codon positions 1 and 2 among Galliformes, while for codon position 3, some species departed from the average composition.
The combined dataset of 12 protein-coding genes (10,886 aligned sites) revealed a single, robust tree using MP and ML, and BI produced a summary of numerous trees (Additional file 4) for the Galliformes. The Megapodiidae was the sister group of the remaining taxa, followed by the Numididae. The remaining clade formed a complex mixture involving the families Tetraonidae, Meleagrididae and Phasianidae. The Phasianidae contained seven lineages, each with very high BSPs as follows: Group 1 contained Arborophila; Group 2 included Tragopan, Lophophorus and Tetraophasis; Group 3 was composed of Chrysolophus, Phasianus, Lophura, and Syrmaticus; Group 4 contained Perdix; Group 5 had Pucrasia; Group 6 held Gallus, Bambusicola and Francolinus; and Group 7 contained Coturnix and Alectoris. A clade containing families Tetraonidae and Meleagrididae branched off from within the Phasianidae. Only the phylogenetic relationships of Pavo and Polyplectron remained unresolved (Additional file 4).
In order to investigate the possibility of a bias owing to substitution saturation, we plotted transitions and transversions against pairwise sequence divergence using 12 mt protein-coding genes (Additional file 5). Codon positions 1 and 2 were not saturated. Codon position 3 was saturated for transitions in the comparison between the Galliformes and outgroups. In addition, some species departed from the average base composition in codon position 3 (Additional file 3). In order to reduce the possible influence of these two biases on codon position 3, we implemented P12 and RY-coding methods. The resulting tree topologies were identical to those of the unweighted schemes. As expected, relatively lower bootstraps values were obtained (Additional file 6). Better resolution and higher BSPs were obtained from DNA datasets that included all substitutions, as opposed to those subjected to weighting.
Both of the combined RNA datasets (tRNAs and rRNAs) obtained less resolution compared to protein-coding genes (Additional file 7 and Additional file 8). The aligned rRNA sequences (combined 12S and 16S rRNA genes) were 2,744 aligned sites in length. The tRNA dataset (combined 22 tRNA genes) contained 1,612 bp of aligned sites. The individual partitions (13 protein-coding genes, 12S rRNA, 16S rRNA and CR) showed very limited power for phylogenetic inference, leaving many unresolved nodes (Additional file 9).
To maximize the amount of phylogenetic information, we pooled all mt genes (protein-coding, RNA and CR) to form a single dataset with a length of 16,508 aligned sites for a genomic-level phylogeny [33]. The BI analysis yielded a topology identical to that produced by the combined 12 protein-coding genes, except for the phylogenetic position of Polyplectron, which did not cluster with the peafowl, but rooted at the base of the Phasianidae, implying non-monophyly of peafowl (Fig. 2).
Regarding the phylogenetic positions of the Cracidae and Odontophoridae, the BI trees based on 10,502 nucleotide positions (Additional file 10) and 3,262 positions (Additional file 10) were very similar to those attained from complete mt genomes (Fig. 2). As expected, some nodes received relatively low statistical support.
Assessing the performance of individual genes
PBS analyses were performed to better understand the contribution of different parts of the mt genome on the genome phylogeny. Among the 16 partitions examined, ND5 provided the greatest contribution to tree resolution, followed by CR, while ATP8 contributed the least (Fig. 3A). In the analyses of PBS values per nucleotide base pair, CR performed the best and ATP8 was the worst (Fig. 3B).
Discussion
The rapid radiation and convergent morphological evolution has confounded the resolution of relationships for many pheasants and partridges. Most previous molecular studies analyzed either one or a few mt genes [9, 12, 13, 18] only or a single nuclear gene [11, 35]. The paucity of data rendered the sequence of cladogenic events, especially within the family Phasianidae, unsolved. In this study, we showed that the number of genes, and thus the quantity of data, proved critical to resolving relationships. Individual genes had limited power to resolve the phylogeny of this group (Additional file 9). In contrast, the complete mt genomes clearly resolved most of the branching order within the pheasants and partridges, with strong nodal support.
At the base of the tree, Alectura lathami was the sister taxon of all other galliforms, followed in sequence by the cracids, new world quails and guineafowls, and then a clade including the Phasianidae, the Tetraonidae and the Meleagrididae (Additional file 10). This branching order was the same as obtained by some studies [9, 12, 18], yet other studies switched the positions of new world quails and guineafowl [13, 17, 20, 36]. Unlike most previous studies based on morphology [15], a single nuclear gene [11], single mitochondrial gene [9], or retrotransposable elements [19], we obtained clear branching orders within the Phasianidae. Arborophila was the sister group to all other phasianids plus the Meleagrididae and Tetraonidae. Whereas the evolutionary relationships of Perdix remained unresolved in previous studies, in our study it clustered with the gallopheasants, and with strong nodal support (Fig. 2; 100% BSP and 1.00 BPP). Tetraophasis clustered independently with Lophophorus and their sister group was Tragopan; this association also enjoyed very high nodal support. And Arborophila did not cluster with other partridges. Therefore, the non-monophyly of the pheasants and partridges was more common than not, including the strongly supported association of Gallus, Bambusicola and Francolinus. Polyplectron did not cluster with Pavo, and Pucrasia did not cluster with other tragopans. These associations revealed that the peafowl and tragopan lineages were not monophyletic.
Our study and that of Kimball and Braun 2008 (Fig. 1c), based on combined data including two mt genes (CytB, ND2) and four nuclear introns (BFib7, DCoH3, OvoG, and Rhod1), were similar in some respects, but have many differences. For example, within Gallus, they resolved the species relationships as (((G. lafayetti, G. varius) G. gallus) G. sonneratti) and our ML and BI analyses resolved them as ((G. gallus, G. varius) (G. lafayetti, G. sonneratti)). Whereas no nodal support was obtained in the former set of relationships, our ML and BI trees were strongly supported at each node (Fig. 2; 100% BSP and 1.00 BPP). Kimball and Braun placed Polyplectron as the sister group to Gallus and Bambusicola but with less than 50% support values, yet all of our analyses located Polyplectron near the base of the clade for the Phasianidae (Fig. 2; 100% BSP and 1.00 BPP). Other discrepancies also occurred, such as the position of Pavo, Perdix and Pucrasia, and all of these differences received high support in our study.
Our study resolved many differences compared to trees based on retrotransposable elements (Fig. 1d) [19, 36]. These studies placed Gallus near the base of the clade for the Phasianidae, followed by Pavo, and the remaining taxa split into two groups: (1) Coturnix and (2) gallopheasants, tragopans, turkey and grouse. In contrast, our analyses did not place Gallus near the base of the Phasianidae, but rather clustered it with Coturnix. This pair then clustered with Pavo, which then became the sister group of gallopheasants, tragopans, turkey and grouse (Fig. 2). Although Kriegs et al. (2007) did not resolve the position of the tragopans, we found them to be the sister group of the gallopheasants, and with very high support (Fig. 2; 100% BSP and 1.00 BPP). Studies of retrotransposable elements resolved the branching orders of seven families of Galliformes, however, many unsolved parallel relationships remained in the Phasianidae (Fig. 1d). Thus, retrotransposable elements were good markers to resolve relationships at the hierarchical levels of family, but seemed to be less powerful at resolving detailed relationships at the hierarchical levels of genus and species. In contrast, mt genomes were very informative at the level of genus/species, due to their relatively rapid rate of mutation.
Relative branch lengths suggest this group has undergone ancient, rapid radiations. Branching order is difficult to resolve during rapid radiations because of insufficient time for numerous genetic and morphological changes to accumulate [9]. Clearly, our combined mt genomes provide a greater abundance of information and thus may have a greater likelihood of fully resolving a tree than individual protein-coding genes and other subsets of mt genes (tRNA, and rRNA).
Because sequencing the complete mt genomes is expensive in both time and resources, the relative performance of individual genes is of great interest. Ranking individual genes by their respective contribution to the total PBS values--a rough indicator of phylogenetic utility--reveals that some genes, such as ND5, CR, ND4, CoxI and 16S, are better indicators of galliform evolution than others (Fig. 3A). Longer genes tend to have more informative sites, and thus a larger total PBS. PBS value per nucleotide base pair can be used to maximize tree-acquisition efficiency. For galliform birds, ND3, ND1, ND2, ND5, ATP6, and ND4 are more informative than other mt genes (Fig. 3B). PBS values can be used with gene length, ease of amplification and sequencing to select a suite of genes for phylogenetic inference, especially given that different suites of genes may provide more information as a function of the tempo of evolution.
Conclusions
Our robust mitogenomic tree indicates that galliform relationships are very complex. The traditional hypothesis of monophyletic lineages of pheasants, partridges, peafowls and tragopans is not supported in this study. Mitogenomics is a powerful tool for resolving the phylogenetic relationships of the Galliformes. Individual mitochondrial genes and nuclear genes seem to be less powerful in resolving phylogenetic relationships within the Galliformes, especially the Phasianidae. Clearly, complete mt genomes can provide more information, and thus are more powerful arbitrators of ambiguous phylogenetic relationships than partial genomes. Therefore, this strategy should serve to further resolve the phylogeny of the Galliformes as more mt genomes are obtained. Given the diversity of species in the Galliformes, and these birds' great complexity in morphological and behavior characters, such as flight, polygamy and sexual dimorphism, this Order can serve as an ideal model for a detailed study of character evolution. Thus, our study not only hypothesizes an evolutionary history of the Galliformes, it also provides primary genetic data for future studies. A caveat of this study is that, our tree tells us a part of story due to the only matrilineal heredity of mtDNA. In particular, our topology conflicts with that derived from nuclear DNA (nuDNA) retrotransposable elements [19, 36]. These conflicts may be due to the different modes of heredity, and different tempos of evolution of mtDNA and nuDNA. Further study is required to resolve the current conflicts.
Methods
Specimens sampling
Muscle or feather tissue was obtained from 14 species. Further, 26 additional complete mt genomes were obtained from GenBank for the Galliformes and Anserformes (Additional file 1). No complete mt genomes were available for representatives of the Cracidae and Odontophoridae. Consequently, we mined GenBank and created a dataset with as many mitochondrial genes as possible for these taxa (Additional file 11).
DNA extraction, PCR amplification, and sequencing
Total genomic DNA was extracted using standard 3-step phenol/chloroform extraction methods [37]. LA-PCR primer sets and segmental amplification primer sets were described previously [38]. An additional 96 species-specific primers were designed (Additional file 12).
LA-PCR amplifications were conducted using the following parameters: initial denaturation at 95°C 4 min, followed by 30 cycles of denaturation at 94°C for 30 sec, 58°C annealing extension for 16 min, and with a final extension at 72°C for 5 min. Subsequently, the LA-PCR products were used for segmental PCR amplification. PCR amplifications were conducted in a 50 μl volume containing 5 μl of 10 × reaction buffer, 0.2 mM dNTPs, 0.2 μM each primer, 1.5 U Taq DNA polymerase (TaKaRa Biosystems), and approximately 10 ng LA-PCR products. PCR amplifications were carried out using the following parameters: 95°C 4 min, 20 cycles of denaturation at 94°C for 1 min, annealing at 60-50°C (1 min; 0.5°C/cycle), extension at 72°C for 1 min, and finally 15 cycles of 94°C 1 min, 50°C 1 min, 72°C 1 min. PCR products were cleaned using Watson RCR Purification Kits (Watson BioTechnologies, Shanghai).
PCR products were sequenced at least three times in both directions on an ABI 3730 Sequencer (Applied Biosystems, Foster, CA, USA) using the ABI PRISM BigDye Terminator v3.0 sequencing kit. DNA sequences were edited using DNAstar Seqman software (DNASTAR Inc., Madison, WI, USA). The newly determined genomes were deposited in GenBank (GenBank accession numbers: FJ752423-FJ752436).
Phylogenetic reconstruction
The sequence data were initially aligned using ClustalX 1.81 [39] with default parameters. Subsequently, the alignment was adjusted manually.
Because compositional bias among species can interfere with tree topology [40–42], prior to phylogenetic reconstruction, we performed tests of stationarity of base composition in TREEPUZZLE 5.2 [43]. Each gene and gene set was tested separately.
The combined sequence datasets of all 12 light-strand-encoded protein coding genes, two rRNA genes and 22 tRNA genes were analyzed separately using maximum likelihood and maximum parsimony (MP) implemented in PAUP* 4.0b10 [44]. MP heuristic searches used tree bisection reconnection (TBR) branch swapping executed for 10000 replicates. Modeltest 3.7 [45] was used to select the preferred models of evolution for ML, under the Akaike Information Criterion [46]. The GTR+I+G model was selected for the 12 coding genes and 22 tRNAs, and the GTR+G model had the best fit for the two rRNA genes. ML heuristic searches used TBR branch swapping executed in 10 replicates with the selected models. Because heuristic searches in PAUP* are very slow, we used two additional fast ML-based inference packages using 1 000 replicates each: RAxML [47] and PHYML [48]. Because their topologies are the same, and only a few bootstrap values are slightly different, we only present trees with bootstrap values from PAUP*.
Bayesian inference (BI) was performed using MrBayes 3.1.2 [49]. The Bayesian posterior probabilities (BPP) used models estimated with Modeltest 3.7 under AIC. Two separate runs were performed with four Markov chains. Each run was conducted with 5 × 106 generations and sampled every 100 generations. When the log-likelihood scores were found to stabilize, a consensus tree was calculated after omitting the first 25% trees as burn-in.
In order to detect the possible bias of substitution saturation, we plotted transitions and transversions against the pairwise sequence divergence using 12 mitochondrial protein-coding genes in DAMBE [50].
Two additional weighting strategies were applied in the analysis of combined 12 protein-coding genes to avoid possible bias of nucleotide composition and saturation: (1) excluding the 3rd codon positions, and (2) recoding the 3rd codon position nucleotides to two-state categories, R (purine) and Y (pyrimidine), i.e., RY-coding. RY-coding can greatly improve consistency in phylogenetic resolution by reducing bias from differences in nucleotide composition [51, 52].
To examine the performance of individual genes, a partitioned Bremer support (PBS) analyses [53] were performed. The 13 protein-coding genes, 12S rRNA, 16S rRNA and control region (CR) partitions were each used to reconstruct the phylogeny using BI.
To maximize the amount of phylogenetic information, the entire mt genome (13 protein-coding genes, two rRNA genes, 22 tRNA genes and CR) was also used to reconstruct the phylogeny using BI, ML and BP methods.
Complete mt genomes were not available for representatives of the Cracidae and Odontophoridae. Thus, we mined GenBank to attain as many mt genes as possible for 11 species in the Cracidae and four in the Odontophoridae (Additional file 11). We combined these data with those from the complete mt genomes. Given the tradeoff between alignment length and taxonomic coverage, we compiled a dataset of 10502 nucleotide sites for 11 species in the Cracidae and 40 in the Galloanserae that have complete mt genomes. In addition, a dataset of 3262 bp was compiled for 11 representatives of the Cracidae, four Odontophoridae, and 40 Galloanserae. In these two datasets, substitution models were estimated with Modeltest 3.7 under AIC, and then BI was carried out.
Additional file 13 gives the evolutionary models, log-likelihood values (-ln L) and settings identified by Modeltest for the different datasets. In PAUP*, all six representatives of the Anseriformes were used as outgroup taxa when calculating the ML and MP trees. Only one taxon can be set as the outgroup in MrBayes. Therefore, we chose Anseranas semipalmata, and subsequently re-rooted the trees to make Anseriformes and Galliformes reciprocally monophyletic sister groups.
References
Cooper MD, Peterson RDA, South MA, Good RA: The functions of the thymus system and the bursa system in the chicken. J Exp Med. 1966, 123: 75-102. 10.1084/jem.123.1.75.
Brown WRA, Hubbard SJ, Tickle C, Wilson SA: The chicken as a model for large-scale analysis of vertebrate gene function. Nat Rev Genet. 2003, 4: 87-98. 10.1038/nrg998.
Pizzari T, Cornwallis CK, Lovlie H, Jakobssen S, Birkhead TR: Sophisticated sperm allocation in male fowl. Nature. 2003, 426: 70-74. 10.1038/nature02004.
Shibusawa M, Nishibori M, Nishida-Umehara C, Tsudzuki M, Masabanda J, Griffin DK, Matsuda Y: Karyotypic evolution in the Galliformes: An examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet Genome Res. 2004, 106: 111-119. 10.1159/000078570.
Stehelin D, Varmus HE, Bishop JM: DNA related to the transforming gene (s) of avian sar-236 VOGT & HU coma viruses is present in normal avian DNA. Nature. 1976, 260: 170-173. 10.1038/260170a0.
von Schantz T, Goeransson G, Andersson G, Froeberg I, Grahn M, Helgee A, Wittzell H: Female choice selects for a viability-based male trait in pheasants. Nature. 1989, 337: 166-169. 10.1038/337166a0.
Fumihito A, Miyake T, Takada M, Ohno S, Kondo N: The genetic link between the chinese bamboo partridge (Bambusicola thoracica) and the chicken and junglefowls of the genus Gallus. Proc Natl Acad Sci USA. 1995, 92: 11053-11056. 10.1073/pnas.92.24.11053.
Kimball RT: Resolution of the phylogenetic position of the Congo peafowl, Afropavo congensis: A biogeographic and evolutionary enigma. Proc Biol Sci. 1997, 264: 1517-1523. 10.1098/rspb.1997.0210.
Kimball RT, Braun EL, Zwartjes PW, Crowe TM, Ligon JD: A molecular phylogeny of the pheasants and partridges suggests that these lineages are not monophyletic. Mol Phylogenet Evol. 1999, 11: 38-54. 10.1006/mpev.1998.0562.
Randi E, Lucchini V, Armijo-Prewitt T, Kimball RT, Braun EL, Ligon JD: Mitochondrial DNA phylogeny and speciation in the tragopans. Auk. 2000, 117: 1003-1015. 10.1642/0004-8038(2000)117[1003:MDPASI]2.0.CO;2.
Armstrong MH, Braun EL, Kimball RT: Phylogenetic utility of avian ovomucoid intron G: A comparison of nuclear and mitochondrial phylogenies in galliformes. Auk. 2001, 118: 799-804. 10.1642/0004-8038(2001)118[0799:PUOAOI]2.0.CO;2.
Kimball RT, Braun EL, Ligon JD, Lucchini V, Randi E: A molecular phylogeny of the peacock-pheasants (Galliformes: Polyplectron spp.) indicates loss and reduction of ornamental traits and display behaviours. Biol J Linn Soc Lond. 2001, 73: 187-198.
Dimcheff DE, Drovetski SV, Mindell DP: Phylogeny of Tetraoninae and other galliform birds using mitochondrial 12S and ND2 genes. Mol Phylogenet Evol. 2002, 24: 203-215. 10.1016/S1055-7903(02)00230-0.
Pereira SL, Baker AJ, Wajntal A: Combined nuclear and mitochondrial DNA sequences resolve generic relationships within the Cracidae (Galliformes, Aves). Syst Biol. 2002, 51: 946-958. 10.1080/10635150290102519.
Dyke GJ, Gulas BE, Crowe TM: Suprageneric relationships of galliform birds (Aves, Galliformes): a cladistic analysis of morphological characters. Zool J Linn Soc. 2003, 137: 227-244. 10.1046/j.1096-3642.2003.00048.x.
van Tuinen M, Dyke GJ: Calibration of galliform molecular clocks using multiple fossils and genetic partitions. Mol Phylogenet Evol. 2004, 30: 74-86. 10.1016/S1055-7903(03)00164-7.
Crowe TM, Bowie RCK, Bloomer P, Mandiwana TG, Hedderson TAJ, Randi E, Pereira SL, Wakeling J: Phylogenetics, biogeography and classification of, and character evolution in, gamebirds (Aves: Galliformes): effects of character exclusion, data partitioning and missing data. Cladistics. 2006, 22: 495-532. 10.1111/j.1096-0031.2006.00120.x.
Pereira SL, Baker AJ: A molecular timescale for galliform birds accounting for uncertainty in time estimates and heterogeneity of rates of DNA substitutions across lineages and sites. Mol Phylogenet Evol. 2006, 38: 499-509. 10.1016/j.ympev.2005.07.007.
Kaiser VB, van Tuinen M, Ellegren H: Insertion events of CR1 retrotransposable elements elucidate the phylogenetic branching order in galliform birds. Mol Biol Evol. 2007, 24: 338-347. 10.1093/molbev/msl164.
Kimball RT, Braun EL: A multigene phylogeny of Galliformes supports a single origin of erectile ability in non-feathered facial traits. J Avian Biol. 2008, 39: 438-445.
Meng Y, Dai B, Ran J, Li J, Yue B: Phylogenetic position of the genus Tetraophasis (Aves, Galliformes, Phasianidae) as inferred from mitochondrial and nuclear sequences. Biochem Syst Ecol. 2008, 36: 626-637. 10.1016/j.bse.2008.01.007.
Johnsgard PA: The pheasants of the world. 1986, Oxford University Press New York
Mindell DP, Thacker CE: Rates of molecular evolution: Phylogenetic issues and applications. Annu Rev Ecol Evol Syst. 1996, 27: 279-303. 10.1146/annurev.ecolsys.27.1.279.
Zardoya R, Meyer A: Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol Biol Evol. 1996, 13: 933-942.
Naylor GJ, Brown WM: Structural biology and phylogenetic estimation. Nature. 1997, 388: 527-528. 10.1038/41460.
Rosenberg MS, Kumar S: Incomplete taxon sampling is not a problem for phylogenetic inference. Proc Natl Acad Sci USA. 2001, 98: 10751-10756. 10.1073/pnas.191248498.
Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al: Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008, 452: 745-749. 10.1038/nature06614.
Slack KE, Jones CM, Ando T, Harrison GL, Fordyce RE, Arnason U, Penny D: Early penguin fossils, plus mitochondrial genomes, calibrate avian evolution. Mol Biol Evol. 2006, 23: 1144-1155. 10.1093/molbev/msj124.
Watanabe M, Nikaido M, Tsuda TT, Kobayashi T, Mindell D, Cao Y, Okada N, Hasegawa M: New candidate species most closely related to penguins. Gene. 2006, 378: 65-73. 10.1016/j.gene.2006.05.003.
Amer SAM, Kumazawa Y: The mitochondrial genome of the lizard Calotes versicolor and a novel gene inversion in South Asian draconine agamids. Mol Biol Evol. 2007, 24: 1330-1339. 10.1093/molbev/msm054.
Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D: Mitochondrial genomes and avian phylogeny: Complex characters and resolvability without explosive radiations. Mol Biol Evol. 2007, 24: 269-280. 10.1093/molbev/msl158.
Kumazawa Y: Mitochondrial genomes from major lizard families suggest their phylogenetic relationships and ancient radiations. Gene. 2007, 388: 19-26. 10.1016/j.gene.2006.09.026.
Yu L, Li YW, Ryder OA, Zhang YP: Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol Biol. 2007, 7: 198-208. 10.1186/1471-2148-7-198.
Desjardins P, Morais R: Sequence and gene organization of the chicken mitochondrial genome: A novel gene order in higher vertebrates. J Mol Biol. 1990, 212: 599-634. 10.1016/0022-2836(90)90225-B.
Smith EJ, Shi L, Tu ZJ: Gallus gallus aggrecan gene-based phylogenetic analysis of selected avian taxonomic groups. Genetica. 2005, 124: 23-32. 10.1007/s10709-004-5184-4.
Kriegs JO, Matzke A, Churakov G, Kuritzin A, Mayr G, Brosius J, Schmitz J: Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes). BMC Evol Biol. 2007, 7: 190-200. 10.1186/1471-2148-7-190.
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Mannual. 1989, New York: Cold Spring Harbor Laboratory Press
Nishibori M, Hayashi T, Tsudzuki M, Yamamoto Y, Yasue H: Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species. Anim Genet. 2001, 32: 380-385. 10.1046/j.1365-2052.2001.00795.x.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The Clustal × windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Versión 1.8. Nucleic Acids Res. 1997, 24: 4876-4882. 10.1093/nar/25.24.4876.
Foster PG, Hickey DA: Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol. 1999, 48: 284-290. 10.1007/PL00006471.
Mooers A, Holmes EC: The evolution of base composition and phylogenetic inference. Trends Ecol Evol. 2000, 15: 365-369. 10.1016/S0169-5347(00)01934-0.
Haddrath O, Baker AJ: Complete mitochondrial DNA geonome sequences of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis. Proc R Soc Lond, B, Biol Sci. 2001, 268: 939-945. 10.1098/rspb.2001.1587.
Schmidt HA, Strimmer K, Vingron M, von Haeseler A: TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. 2002, Oxford Univ Press, 18: 502-504.
Swofford DL: PAUP* 4.06. 10: Phylogenetic analysis using parsimony. 2003, Sunderland (MA): Sinauer Associates
Posada D, Buckley TR: Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004, 53: 793-808. 10.1080/10635150490522304.
Shepherd PR, Salvesen G, Toker A, Ladbury J, Mayo KH, Baici A, Board P, Booth P, Bourne Y, Doyle D: The molecular mechanism for the spectral shifts between vertebrate ultraviolet-and violet-sensitive cone visual pigments. Biochem J. 2002, 367: 129-135. 10.1042/BJ20020483.
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Guindon S, Gascuel O: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003, 52: 696-704. 10.1080/10635150390235520.
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003, 19: 1572-1574. 10.1093/bioinformatics/btg180.
Xia X, Xie Z: DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001, 92: 371-373. 10.1093/jhered/92.4.371.
Phillips MJ, Penny D: The root of the mammalian tree inferred from whole mitochondrial genomes. Mol Phylogenet Evol. 2003, 28: 171-185. 10.1016/S1055-7903(03)00057-5.
Harrison GL, McLenachan PA, Phillips MJ, Slack KE, Cooper A, Penny D: Four new avian mitochondrial genomes help get to basic evolutionary questions in the late Cretaceous. Mol Biol Evol. 2004, 21: 974-983. 10.1093/molbev/msh065.
Baker RH, DeSalle R: Multiple sources of character information and the phylogeny of Hawaiian drosophilids. Syst Biol. 1997, 46: 654-673.
Acknowledgements
Special thanks to Christopher Blair and Mumtaz Baig for their suggestions. This work was supported by National Basic Research Program of China (973 Program, 2007CB411600), National Natural Science Foundation of China (30621092), and Bureau of Science and Technology of Yunnan Province.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
YYS: contributed to data analysis and manuscript writing; LL: carried out the experiment work; YBS: carried out the experiment work; BSY: contributed to data analysis; XJY: contributed to data analysis; RWM: contributed to data analysis and manuscript writing; YPZ: designed the study and prepared the manuscript; All authors have read and approved the final manuscript.
Electronic supplementary material
12862_2010_1348_MOESM1_ESM.DOC
Additional file 1: Source of sequence data. Source of sequence data for mitochondrial genomes and general characteristics of 40 species in the Galloanserae. (DOC 91 KB)
12862_2010_1348_MOESM2_ESM.XLS
Additional file 2: Test of stationarity of base composition. Test of stationarity of base composition in TREEPUZZLE 5.0. The chi-square test compares the nucleotide composition of each sequence to the frequency distribution assumed in the maximum likelihood model. (XLS 178 KB)
12862_2010_1348_MOESM3_ESM.TIFF
Additional file 3: Base composition for the 12 protein-coding gene set. (A) All codon sites; (B) 1st codon position; (C) 2nd codon position; (D) 3rd codon position. (TIFF 7 MB)
12862_2010_1348_MOESM4_ESM.PDF
Additional file 4: Bayesian tree based on 12 mitochondrial protein-coding genes. In order to emphasize the topology of Galliformes, we did not include the outgroup on the tree. Numbers are maximum likelihood bootstrap support and Bayesian posterior probabilities. Branches are drawn proportionally to the average number of expected DNA substitutions per site among all trees sampled after a burn-in period, as indicated by the scale at the bottom left. Species belonging to the Tribe Phasianini are marked in purple, and to Tribe Perdicini in green. (PDF 31 KB)
12862_2010_1348_MOESM5_ESM.PDF
Additional file 5: Substitution saturation of 12 mitochondrial protein-coding genes. Transitions and transversions plotted against the pairwise sequence divergence for 12 mitochondrial protein-coding genes. (A) 1st codon position; (B) 2nd codon position; (C) 3rd codon position; (D) all codon sites. (PDF 84 KB)
12862_2010_1348_MOESM6_ESM.PDF
Additional file 6: Bayesian phylogenetic analyses of RY-coding and exclusion the 3rd codon position. Bayesian phylogenetic analyses of two weighting strategies in the combined 12 protein-coding gene sets. Bayesian posterior probabilities >70% are indicated on the branches. (A) Recoding the 3rd codon position nucleotides as to two-state categories, R (purine) and Y (pyrimidine), (RY-coding); (B) Excluding the 3rd codon position. (PDF 25 KB)
12862_2010_1348_MOESM7_ESM.PDF
Additional file 7: Bayesian tree for 22 tRNA genes. Bayesian inference consensus tree for the Galliformes based on combined data from mitochondrial 22 tRNA genes. Anseriformes forms the outgroup. Bayesian posterior probabilities >70%, and maximum likelihood bootstrap proportions >50% are indicated on the branches. (PDF 17 KB)
12862_2010_1348_MOESM8_ESM.PDF
Additional file 8: Bayesian tree based on combined data from 12S rRNA and 16S rRNA genes. Bayesian inference consensus tree for Galliformes based on combined data from mitochondrial 12S rRNA and 16S rRNA genes. Anseriformes forms the outgroup. The Bayesian posterior probabilities >70%, and maximum likelihood bootstrap proportions >50% are indicated on the branches. (PDF 17 KB)
12862_2010_1348_MOESM9_ESM.PDF
Additional file 9: Bayesian analyses of individual mt genes. Bayesian inference analyses of individual mt genes and control region (CR). Each run was conducted with 5,000,000 generations and sampled every 100 generations. Bayesian Posterior Probabilities >70% are indicated on the branches. (A) ND1, 972 aligned sites; (B) ND2, 1,038 aligned sites; (C) ND3, 348 aligned sites; (D) ND4L, 291 aligned sites; (E) ND4, 1,377 aligned sites; (F) ND5, 1,818 aligned sites; (G) ND6, 519 aligned sites; (H) CoxI, 1,548 aligned sites; (I) CoxII, 681 aligned sites; (J) CoxIII, 783 aligned sites; (K) ATP6, 681 aligned sites; (L) ATP8, 165 aligned sites; (M) CytB, 1,137 aligned sites; (N) CR, 1,294 aligned sites; (O) 12S, 1,047 aligned sites; (P) 16S, 1,695 aligned sites. (PDF 223 KB)
12862_2010_1348_MOESM10_ESM.PDF
Additional file 10: Bayesian trees based on combined datasets of the Cracidae and Odontophoridae. Bayesian inference (BI) consensus trees based on combined datasets of the Cracidae and Odontophoridae. Anseriformes forms the outgroup. Bayesian posterior probabilities > 70% are indicated on the branches. (A) The BI tree for the dataset of 10,502 nucleotide positions for 11 species in the Cracidae (marked in red) and 40 galliform/anseriform birds that have complete mt genomes; (B) The BI tree for the dataset of 3,262 aligned nucleotide positions for 11 species in the Cracidae (marked in red), four in the Odontophoridae (marked in blue) and 40 galliform/anseriform birds. (PDF 85 KB)
12862_2010_1348_MOESM11_ESM.XLS
Additional file 11: Source of sequence data for the Cracidae and Odontophoridae. No complete mitochondrial genomes were available for the Cracidae and Odontophoridae in GenBank. Thus, we mined GenBank and created a dataset with as many genes as possible for them. (XLS 29 KB)
12862_2010_1348_MOESM12_ESM.XLS
Additional file 12: Primers for amplifying the complete mitogenomes. List of primers used in this mitogenomic study of the Galliformes. (XLS 47 KB)
12862_2010_1348_MOESM13_ESM.DOC
Additional file 13: Parameters of evolutionary models. Evolutionary models, log-likelihood values (-ln L), and settings identified by Modeltest for different DNA sequence datasets from the Galiformes. (DOC 57 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Shen, YY., Liang, L., Sun, YB. et al. A mitogenomic perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae. BMC Evol Biol 10, 132 (2010). https://doi.org/10.1186/1471-2148-10-132
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-10-132