Abstract
Background
Nuclear DNA sequences provide genetic information that complements studies using mitochondrial DNA. Some 'universal' primer sets have been developed that target introns within protein-coding loci, but many simultaneously amplify introns from paralogous loci. Refining existing primer sets to target a single locus could circumvent this problem.
Results
Aldolase intron 'G' was amplified from four fish species using previously described primer sets that target several loci indiscriminately. Phylogenetic analyses were used to group these fragments and other full-length aldolase proteins from teleost fishes into orthologous clades and a primer set was designed to target specifically an intron within the aldolase-B locus in acanthopterygian fishes. DNA amplifications were tried in a variety of acanthopterygian fishes and amplification products, identifiable as aldolase-B intron 'G', were observed in all atherinomorph and percomorph taxa examined. Sequence variation within this locus was found within and among several species examined.
Conclusions
Using 'universal' primer sets coupled with phylogenetic analyses it was possible to develop a genetic assay to target a specific locus in a variety of fish taxa. Sequence variation was observed within and among species suggesting that this targeted assay might facilitate interspecific and intraspecific comparisons.
Similar content being viewed by others
Background
DNA sequences from the nuclear genome provide genetic information that complements and extends intra- and inter-specific studies using mitochondrial DNA [e.g., [1, 2]]. Sets of oligonucleotide primers have been developed that target introns within nuclear-encoded loci [e.g., [3–5]]; however, some 'universal' primer sets amplify introns from several loci simultaneously [5]. Although each locus would provide an independent estimate of phylogenetic relationship, the amplification of multiple loci might cause confusion regarding relationships among loci across taxa. Developing primer sets that target single loci [6, 7] or refining existing primer sets that amplify multiple loci to target a single locus could circumvent this problem. We demonstrate the latter approach by redesigning 'universal' primers to target the aldolase-B (Ald-B) locus in a sample of acanthopterygian fishes.
Results and discussion
Phylogenetic analyses strongly supported a grouping of ALD-B proteins from Salmo salar[8], Sparus auratus[9] and partial amino acid sequences isolated from Micropterus and Lepomis (Fig. 1B). Similarly, a moderately supported clade contained ALD-C proteins from Carassius auratus[10], Sphoeroides nephelus (T. Z. Berardini and D. R. Tolan, unpublished GenBank submission) and partial amino acid sequences inferred from Micropterus and Elassoma (Fig 1B). A third class of aldolase isolated from Morone clustered outside of the ALD-C clade (Fig. 1B); since most vertebrates express three aldolase isozymes (A, B and C; [11]), we presume this clone represents ALD-A.
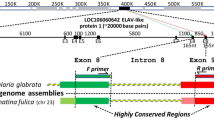
A. Alignment of aldolase fragments from teleost fishes; intron sequences have been removed from starred sequences to clarify diagnostic positions and the overall alignment. Numbers in brackets are GenBank [16] accession codes, circles mark clones of Aldl-5'/Ald2-3' amplifications, and stars mark direct sequences from AldBF2/Ald2-3' genomic DNA amplifications. Numbers above sequences are amino acid positions in Sparus aldolase-B [9] and mark the 3' residue used to design the primers in Figure 2. Position 186 is the 3' residue of primer Ald2-3' [3]. Periods indicate identical amino acid residue to that of the reference taxon; dashes are missing data. The arrow indicates the position of intron 'G'. Question marks after locus designations indicate inferred orthologies from Figure 1B. B. UPGMA dendrogram relating teleost aldolase sequences from this study and GenBank [16]. Numbers are bootstrap support (1000 replicates) for the indicated node.
Methods outlined in Quattro and Jones [6] were used to identify unique amino acid residues shared among ALD-B that differed from ALD-A and ALD-C. Sequences identified as ALD-B are characterized by a cysteine residue (position 143 in the Sparus sequence) not found in ALD-A or ALD-C (Fig. 1A). Similarly, all ALD-B sequences contain a cysteine residue at position 158 that is occupied by a threonine in ALD-A and ALD-C (Fig. 1A). Primers were designed with these diagnostic amino acid positions at the 3' terminus (Fig. 2):
AldBF: 5' – TGC GCC CAG TAC AAG AAG GAC GGT TG – 3'
AldBF2: 5' – CTC AAG ATC TCG GAC GGC TG – 3'
AldBF and AldBF2 were paired with the original Ald2-3' oligonucleotide [3] to target the 'G' intron of the Ald-B locus. DNA amplifications were tried from a variety of acanthopterygian fishes using the PCR conditions described above. Amplifications with the AldBF or AldBF2 primer were often in quantities insufficient for direct sequencing; re-amplifications were performed with small aliquots (1 μl of a 1/200 dilution) of the AldBF/Ald2-3' PCR products, AldBF2/Ald2-3' oligonucleotides, and annealing temperatures of 54°C to 60°C. Single amplification products were observed in all atherinomorph (Gambusia holbrooki, Fundulus heteroclitus, Cyprinodon variegatus, Poeciliopsis lucida) and percomorph (Astronotus ocellatus, Cichlasoma octofasciatum, E. evergladei, Etheostoma saludae, M. americana, M. saxatilis, Paralichthys dentatus) taxa examined (Fig. 1A). Individual PCR products were sequenced following the method of Salminen [12] and translated into protein to determine orthology. We conclude that amplifications were specific for the Ald-B locus based on the following evidence:
1. The AldBF and AldBF2 primers were designed so that locus-specific nucleotides occupied the extreme 3' portions of the oligonucleotide, thus these primers should be highly specialized for the Ald-B locus.
2. All amplification products from genomic DNA using the AldBF and AldBF2 primers contained amino acid residues diagnostic for other teleost ALD-B at positions 161 (alanine in all but S. salar), 165 (alanine in all but P. dentatus), and/or 183 (leucine) (Fig. 1A).
3. We did not observe multiple bands at any nucleotide position on sequencing gels as expected if multiple loci were amplified during PCR.
4. Sequencing of cloned amplification products yielded homogeneous sequences with amino acid positions diagnosable as unique to teleost ALD-B.
Although intron position was conserved, intron size was variable and ranged from 70 – 100 base pairs (bp) across taxa (Fig. 1A shows only coding region with the intervening intron removed so that orthology is presented more clearly). Sequence variation within the Ald-B intron was found within and among several species examined. For example, five of 86 (5.8%) positions differentiate M. americana from M. saxatilis, and a number of substitutions separate species of pygmy sunfishes (Genus Elassoma) and darters (Genus Etheostoma), thus these sequences might be of use in interspecific phylogenetic and/or forensic applications (J. M. Quattro, unpublished data). Similarly, intraspecific variation has been observed within a small sample of P. dentatus and among populations of E. collis, and many of the Ald-B introns assayed contained microsatellites including all surveyed species of Elassoma.
Conclusions
Nucleotide primers were designed to target specifically an intron within the aldolase-B locus in acanthopterygian fishes. DNA amplifications were tried in a variety of acanthopterygian fishes and amplification products, identifiable as aldolase-B intron 'G', were observed in all atherinomorph and percomorph taxa examined. Intron size was variable across taxa and sequence variation was found within and among several species examined suggesting this locus might be useful for intraspecific and interspecific analyses. Our sampling of taxa is biased towards those on which our lab currently works; we have not fully explored the taxonomic breadth over which these primer sets might be useful. However, the Ald-B locus described here, along with other single-copy loci [6, 7], might prove useful as markers for studies of intra- and inter-specific phylogeny in many acanthopterygian fishes.
Materials and methods
Aldolase intron 'G' was amplified from four fish species (Elassoma okefenokee, Lepomis macrochirus, Micropterus salmoides and Morone americana) using the primer set Ald1-5' and Ald2-3' [3]. Template DNA was extracted from skeletal muscle using the QIAmp Tissue Kit (Qiagen); concentration was estimated by comparison to standards using the intensity of ethidium bromide fluorescence. Each 50 μl PCR contained: 10 mM Tris-HCL (pH 8.3), 2.5 mM MgCl2, 50 mM KC1, 0.01% NP-40, 0.01% Triton X-100, 5% DMSO, 200 μM each dNTP, 10 pmol primer, 100 ng template DNA, and 2 U Promega Taq DNA polymerase. Reactions were cycled in a MJ Research PTC-100 programmable thermal controller under the following conditions: an initial step of 4 min at 94°C, followed by 40 cycles of 1 min at 94°C, 1 min at 48°C, 2 min at 72°C. Upon electrophoresis in 1.2% agarose and visualization by ethidium bromide staining, multiple amplification products were evident in reactions with L. macrochirus and M. salmoides template, whereas only single products were noticeable in reactions with E. okefenokee and M. americana.
Amplification products were cloned in T-vector (Promega) and sequenced manually (Sequenase, version 2, USB). The starting and end positions of the introns were determined by identification of consensus splice sites (GT and AG, respectively; [13]). mtron sequences were removed to yield small portions of coding region (51 amino acids; Fig. 1A). Phylogenetic analyses were used to group these fragments and other full-length aldolase proteins from teleost fishes into orthologous clades (Fig. 1B). UPGMA trees were constructed with MEGA (version 1.0; [14]) using p-distances and the pair-wise deletion option for missing data. Bootstrapping [15] was used as a measure of clade stability.
References
Karl SA, Avise JC: PCR-based assays of Mendelian polymorphisms from anonymous single-copy nuclear DNA: Techniques and applications for population genetics. Mol Biol Evol. 1993, 10: 342-361.
Slade RW, Moritz C, Heideman A: Multiple nuclear-gene phylogenies: Application to pinnipeds and comparison with a mitochondrial DNA gene phylogeny. Mol Biol Evol. 1994, 11: 341-356.
Lessa EP, Applebaum G: Screening techniques for detecting allelic variation in DNA sequences. Mol Ecol. 1993, 2: 119-129.
Palumbi SR, Baker CS: Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol Biol Evol. 1994, 11: 426-435.
Palumbi SR: Nucleic acids II: the polymerase chain reaction. In: Molecular Systematics, Second Edition (Edited by Hillis DM, Moritz C, Mable BK) Sunderland, Massachusetts Sinauer. 1996, 205-247.
Quattro JM, Jones WJ: Amplification primers that target locus-specific introns in Actinopterygian fishes. Copeia. 1999, 1999: 191-196.
Quattro JM, Jones WJ, Grady JM, Rohde FC: Gene-Gene Concordance and the Phylogenetic Relationships Among Rare and Widespread Pygmy Sunfishes (Genus Elassoma). Mol Phy Evol. 2001, 18: 217-226. 10.1006/mpev.2000.0884.
Llewellyn L, Sweeney GE, Ramsum VP, Rogers SA, Wigham T: Cloning and unusual expression profile of the aldolase B gene from Atlantic salmon. Biochim Biophys Ada. 1998, 1443: 375-380. 10.1016/S0167-4781(98)00229-2.
Llewellyn L, Ramsum VP, Sweeney GE, Wigham T, Santos CR, Power DM: Cloning and characterization of a fish aldolase B gene. Biochim Biophys Ada. 1995, 1263: 75-78. 10.1016/0167-4781(95)00096-Y.
Berardini TZ, Drygas-Williams M, Callard GV, Tolan DR: Identification of neuronal isozyme specific residues by comparison of goldfish aldolase C to other aldolases. Comp Biochem Physiol A. 1997, 117: 471-476. 10.1016/S0300-9629(96)00396-9.
Zhang R, Yatsuki H, Kusakabe Y, Iwabe N, Miyata T, Imai T, Yoshida M, Hori K: Structures of cDNAs encoding the muscle-type and non-muscle-type isozymes of lamprey fructose biphosphate aldolases and the evolution of aldolase genes. J Biochem. 1995, 117: 545-553.
Salminen M: Rapid and simple solid phase direct sequencing of in vivo HIV-1 quasispecies. AIDS Res Hum Retroviruses. 1992, 8: 1733-1742.
Mount SM: A catalogue of splice junction sequences. Nucleic Acids Res. 1982, 10: 459-472.
Kumar S, Tamura K, Nei M: MEGA: Molecular Evolutionary Genetics Analysis. Version 1.0. The Pennsylvania State University, University Park, PA. 1993
Felsenstein J: Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985, 39: 783-791.
Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF: GenBank. Nucleic Acids Res. 1998, 26: 1-7. 10.1093/nar/26.1.1.
Acknowledgements
The authors thank J. Grady, T. Greig, T. Merritt, and J. Staton for constructive comments on the manuscript, and L. Shi and P. Owen for technical support. Research was sponsored by the Cooperative Institute for Fisheries Molecular Biology (FISHTEC; NOAA/NMFS (RT/F-1)), the Venture Fund and Marine Science Program, University of South Carolina, and NSF grant OCE-9814172.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Quattro, J.M., Jones, W.J. & Oswald, K.J. PCR primers for an aldolase-B intron in acanthopterygian fishes. BMC Evol Biol 1, 9 (2001). https://doi.org/10.1186/1471-2148-1-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-1-9