Abstract
Background
Notch signaling is highly conserved in the metazoa and is critical for many cell fate decisions. Notch activation occurs following ligand binding to Notch extracellular domain. In vitro binding assays have identified epidermal growth factor (EGF) repeats 11 and 12 as the ligand binding domain of Drosophila Notch. Here we show that an internal deletion in mouse Notch1 of EGF repeats 8–12, including the putative ligand binding domain (lbd), is an inactivating mutation in vivo. We also show that maternal and zygotic Notch1lbd/lbdmutant embryos develop through gastrulation to mid-gestation.
Results
Notch1lbd/lbdembryos died at mid-gestation with a phenotype indistinguishable from Notch1 null mutants. In embryonic stem (ES) cells, Notch1lbd was expressed on the cell surface at levels equivalent to wild type Notch1, but Delta1 binding was reduced to the same level as in Notch1 null cells. In an ES cell co-culture assay, Notch signaling induced by Jagged1 or Delta1 was reduced to a similar level in Notch1lbdand Notch1 null cells. However, the Notch1lbd/lbdallele was expressed similarly to wild type Notch1 in Notch1lbd/lbdES cells and embryos at E8.75, indicating that Notch1 signaling is not essential for the Notch1 gene to be expressed. In addition, maternal and zygotic Notch1 mutant blastocysts developed through gastrulation.
Conclusion
Mouse Notch1 lacking the ligand binding domain is expressed at the cell surface but does not signal in response to the canonical Notch ligands Delta1 and Jagged1. Homozygous Notch1lbd/lbdmutant embryos die at ~E10 similar to Notch1 null embryos. While Notch1 is expressed in oocytes and blastocysts, Notch1 signaling via canonical ligands is dispensable during oogenesis, blastogenesis, implantation and gastrulation.
Similar content being viewed by others
Background
Notch1 is a heterodimeric, type I transmembrane receptor that is required for cell fate decisions throughout the metazoa [1, 2]. The Notch1 extracellular domain contains 36 tandem epidermal growth factor-like (EGF) repeats, and three Lin/Notch repeats. Of the 36 EGF repeats in Drosophila Notch, deletion of only EGF repeats 11 and 12 prohibits the binding of the Notch ligands Delta and Serrate in in vitro binding assays [3, 4]. Notch signaling in mammals is also initiated by binding to canonical Notch ligands (Delta and Jagged) on adjacent cells. Ligand binding activates Notch signaling through two proteolytic cleavage events, first in the extracellular domain by the ADAM10 metalloprotease [5], and subsequently in the transmembrane domain by a presenilin complex with γ-secretase activity [6, 7]. The released Notch intracellular domain (NICD) translocates to the nucleus and binds to the CSL (CBF1, Suppressor of hairless, Lag-1) transcriptional repressor [6]. The NICD/CSL complex recruits co-activators including mastermind (MAML), and up-regulates a number of target genes including the HES (Hairy/Enhancer of Split) family of basic helix-loop-helix transcriptional regulators.
The Notch1 gene has been inactivated in mice by inserting a neomycin gene into EGF32 (Notch1in32; [8]) or by deleting a large internal fragment from aa 1056–2049 that spans the transmembrane domain (Notch1tm1/Con/1; [9]). The Notch1in32 mutation generates a null allele [10] and Notch1tm1/Con/1 homozygotes have an indistinguishable embryonic lethal phenotype. Notch1 null embryos die at mid-gestation around E10, with severe defects in somitogenesis, neurogenesis, vasculogenesis and cardiogenesis. The phenotype of flies expressing Notch with the ligand binding domain deletion is not known. Thus in order to investigate biological consequences of this type of Notch mutation, we generated a mouse Notch1 mutation termed Notch1lbdby deleting EGF repeats 8–12 (aa 290–481), which include the putative Notch1 ligand binding domain. We show that Notch1lbd is expressed on the cell surface but cannot bind to canonical Notch ligands nor signal in response to these ligands. Homozygous Notch1lbd/lbdembryos exhibit defects during embryogenesis similar to Notch1 null mutants. However, Notch1lbd transcripts are expressed at levels similar to wild type in ES cells and in E8.75 Notch1lbd/lbdembryos, indicating that canonical Notch1 signaling is not essential for Notch1 gene expression during early embryogenesis. In addition, while Notch1 is expressed in oocytes and blastocysts [11, 12], we show that oocyte-specific inactivation of Notch1 does not affect oogenesis or fertilization and that maternal and zygotic mutants proceed normally through blastogenesis, implantation and gastrulation.
Results
Notch signaling defects in Notch1lbd/lbdembryos
To generate mice with Notch1 lacking the putative ligand binding domain, embryonic stem (ES) cells with loxP sequences flanking exons 6 – 8 of mouse Notch1 were generated by gene targeting (Fig. 1A; [13]). Exons 7 and 8 encode EGF11 and EGF12 and exon 6 was included in order that the mutant Notch1 was ~20 kDa lower in molecular weight. Two independent ES colonies selected for resistance to G418 were shown by Southern analysis to carry a targeted Notch1 allele (Fig. 1B). Chimeric mice carrying the mutant allele were crossed with mice expressing the MeuCre40 recombinase transgene [14] to obtain mice with a Notch1lbdallele after deletion of exons 6 – 8 along with the HSVtk/Neo cassette (Fig. 1A). Southern blot and PCR analysis of genomic DNA were used to genotype E9.5 embryos of Notch1+/lbdcrosses (Fig. 1C). All expected genotypes were represented at this stage. However, only wild type and heterozygous pups were born from 6 litters (Table 1).
Targeting of the Notch1 gene. (A) Schematic representation of the floxed region of the mouse Notch1 gene. Exons 6 – 8 (* designates a T466A point mutation termed Notch112f described by [13]) and the HSVTk/Neo cassette were removed by Cre recombinase to generate the Notch1lbdallele. The diagram of Notch1 ECD shows EGF repeats as rectangles and LIN repeats as ovals. The ligand binding domain in EGF repeats 11 and 12 is striped. Amino acids 290–481 were removed by the Notch1lbdmutation. PCR primers 5F, 6R and 9R and the P1415 probe are indicated. B1: BamHI; E1: EcoRI; H3: HindIII. (B) Southern blot analysis of two targeted ES clones (C32 and 132D) by hybridization with probe P1415 after digestion with HindIII or HindIII and EcoRI. (C) Southern blot analysis and PCR genotyping of yolk sac genomic DNA from E9.5 embryos from a Notch1+/lbdheterozygous cross. Genomic DNA was digested with BamHI and probed with P1415. (D) RT-PCR analysis of total RNA from ES cells of the genotypes shown. A hybrid band was obtained from Notch1+/lbdcDNA. (E) E9.5 embryos exhibited defective vascularization of yolk sac and retarded development of Notch1lbd/lbdembryos, but no apparent differences between wild type and heterozygous progeny.
To determine when Notch1lbd/lbdembryos die, embryos from Notch1+/lbdcrosses were examined during embryogenesis. Notch1lbd/lbdembryos were indistinguishable from wild type at ~E8.75, but by ~E9.5 Notch1lbd/lbdembryos were severely growth-retarded, with a tube-like heart, distended pericardial sac, and defective vascularization of the yolk sac (Fig. 1D). By ~E10.5, many mutant embryos were resorbed and all mutant embryos were resorbed by ~E11.5. Therefore Notch1lbd/lbdembryos exhibited global defects in Notch signaling with a phenotype indistinguishable from Notch1in32 [8] or Notch1tm1/Con/1 [9] null embryos.
Notch1lbdis expressed at the cell surface but does not signal
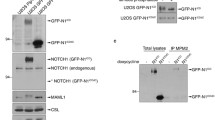
Blastocysts from heterozygous Notch1+/lbdcrosses were used to isolate embryonic stem (ES) cell lines of each genotype. Reverse transcription (RT)-PCR of total RNA showed that Notch1lbd/lbdES cells expressed Notch1 transcripts at levels similar to wild type and heterozygous ES cells (Fig. 1E). Notch1lbd/lbdES cells had a similar growth rate to Notch1+/lbdand Notch1+/+ ES cells (Fig. 2A). This was also observed with Notch1 null ES cells [10]. An antibody to the extracellular domain of Notch1 (8G10) detected the ~300 kD full length Notch1 in wild type ES cells and the ~280 kDa truncated Notch1 in Notch1lbd/lbdES cells (Fig. 2B). The ~180 kDa Notch1 extracellular domain was not routinely observed, but when present it was in similar amounts in Notch1lbd/lbdand Notch1+/+cells. Flow cytometry showed that equivalent amounts of wild type and mutant Notch1 receptors were present on the surface of Notch1+/+ and Notch1lbd/lbdES cells, respectively (Fig. 2C). Therefore the internal deletion that includes the putative ligand binding domain did not affect Notch1 stability or trafficking to the cell surface.
Notch1 lacking the ligand binding domain is expressed on the cell surface but does not signal. (A) Growth curves of ES cells isolated from E3.5 Notch1+/+, Notch1+/lbdand Notch1lbd/lbdblastocysts. Bars represent mean ± SD. (B) Western blot analysis of ES cell lysates (50 μg protein). Full length Notch1 was detected by antibody 8G10 and activated Notch1 was detected by antibody Val1744. Blots were stripped and reprobed using anti-β-Tubulin III. Data are representative of 3 experiments. (C) Flow cytometry of cell surface Notch1 in Notch1lbd/lbdand Notch1+/+ ES cells using anti-Notch1 ECD antibody 8G10 followed by Alexa-488 conjugated anti-hamster IgG. Grey profiles are secondary antibody alone. Profiles are representative of two experiments. (D) Notch ligand binding. ES cells were incubated with soluble Delta1-Fc followed by PE-conjugated anti-human IgG and analyzed by flow cytometry. Notch1in32/in32 null ES cells were line 290-2. EDTA in the binding buffer inhibited binding to all Notch receptors. Bars represent mean ± SEM; n = 5 for Notch1+/+ and Notch1lbd/lbd; n = 3 for Notch1in32/in32 ES cells. (E) ES cells were assayed for Notch signaling after transfection of the Notch TP-1 reporter construct by co-culturing with L cells expressing Delta1 or Jagged1 compared to control L cells. Bars represent fold-activation ± SEM for Notch1+/+ (white), Notch1lbd/lbdand Notch1in32/in32 (gray) (n = 4; ** P < 0.01, *** P < 0.001).
When Notch1 binds canonical Notch ligands, it undergoes cleavage by γ-secretase and the new N-terminus of activated Notch1 may be detected by the antibody Val1744 [15]. Western blot analysis revealed a robust signal for activated Notch1 in cultured wild type ES cells but no corresponding signal was observed in Notch1lbd/lbdES cells (Fig. 2B). Thus while Notch1lbd is expressed at the cell surface it is not activated under conditions that activate wild type Notch1, presumably because of the loss of its ligand binding domain. Indeed the Notch ligand Delta1 had reduced binding to Notch1lbd/lbdES cells (Fig. 2D). ES cells express each of the four mammalian Notch receptors and all would be expected to bind Delta1. To confirm that the reduced binding of Delta1 to Notch1lbd/lbdis due to the Notch1lbdmutation, we examined Delta1 binding to Notch1 null ES cells (Notch1in32/in32) termed 290-2 which lack Notch1 on the cell surface [10]. The binding of Delta1 to 290-2 cells was reduced to the same extent as to Notch1lbd/lbdcells (Fig. 2D). Therefore, the deletion of EGF repeats 8–12 in mouse Notch1 eliminates Delta1 binding to Notch1 as expected, but binding to other Notch receptors remains. This residual binding was prevented by including EDTA in the binding buffer under conditions that prevent Notch/ligand binding but do not release Notch receptors from the cell surface. Delta1-induced Notch1 signaling was also reduced in Notch1lbd/lbdES cells as shown by a co-culture reporter assay which detects signaling through all four Notch receptors (Fig. 2E). A second canonical Notch ligand, Jagged1, was also defective at inducing Notch signaling in Notch1lbd/lbdES cells (Fig. 2E). Delta1- and Jagged1-induced Notch signaling was also reduced, but not eliminated, in Notch1in32/in32 ES cells which are Notch1 null [10] (Fig. 2E). Residual Notch signaling presumably reflects the presence of the other three Notch receptors.
Notch1 signaling is not essential for Notch1gene expression in early embryogenesis
The Notch1lbdmutant allele is transcribed similarly to the wild type allele in Notch1lbd/lbdES cells (Fig. 1E) but Notch1lbd/lbdES cells are defective in Notch1 signaling (Figs. 1 and 2). The Notch1lbdmutant allele therefore allowed us to determine if signaling via Notch1 is required for the Notch1 gene to be expressed in vivo. Notch1+/lbdfemales were crossed with Notch1+/lbdmales and embryos were examined at mid-gestation. Notch1lbd/lbdembryos were morphologically indistinguishable from wild type embryos at E8.75 and were examined for Notch1 gene expression by in situ hybridization. It can be seen in Fig. 3A that Notch1 was expressed in the presomitic mesoderm (PSM) the forming somite (arrows) and the neural tube. The hybridization signal obtained for wild type and Notch1lbd/lbdembryos was similar. Therefore expression of the Notch1 gene was not markedly altered by the loss of canonical Notch1 signaling in early embryogenesis. By contrast, following removal of Pofut1 [16] or Mind bomb 1 [17, 18], both of which inhibit Notch signaling through all four Notch receptors, Notch1 expression is reduced in the PSM and enhanced in neural tube and mesencephalon of E8.75 embryos. At E9.0, Notch1 transcripts were markedly reduced in Notch1lbd/lbdembryos (Fig. 3B). By E9.5 when Notch1lbd/lbdembryos were dying, Notch1 gene expression was severely reduced (Fig. 3C).
Whole mount in situ hybridization of Notch1 pathway and somitogenic genes. Control (Notch1+/+) embryos (left) and mutant Notch1lbd/lbddenoted lbd/lbd (right) embryos were probed together. Arrows point to highest expression. (A) Notch1 expression in E8.75 control and Notch1lbd/lbdembryos was similar (~9.7 kb probe). (B) At E9.0 Notch1 expression was reduced in Notch1lbd/lbdcompared to control embryos (~4.7 kb probe). (C) At E9.5 Notch1 expression was barely detectable in Notch1lbd/lbdembryos (~9.7 kb probe). (D) Expression of the Notch target gene Hes5 was reduced in mutant embryos at E9.5, with residual expression in brain. (E) Myogenin was poorly and diffusely expressed in E9.5 Notch1lbd/lbdembryos. (F) Uncx4.1 was expressed in the caudal compartment of formed somites of control but was missing from the somitic region of E9.5 Notch1lbd/lbdembryos. Uncx4.1 was up-regulated in the midbrain (asterisk) of E9.5 Notch1lbd/lbdembryos (n ≥ 3 for mutant embryos for each probe).
Whole mount in situ hybridization provided additional confirmation that the Notch1lbdmutation inactivates Notch1 signaling. The Notch1 target and somitogenic genes Hes5, Myogenin and Uncx4.1 were examined in embryos at E9.5. At that stage Hes5 is expressed in neural tube, brain and the forming and formed somites. The Hes5 gene is a known target of Notch1 signaling and its expression was severely reduced in Notch1lbd/lbdembryos (Fig. 3D). Myogenin is expressed in mature somites of wild type embryos at E9.5. In Notch1lbd/lbdembryos which had 13–17 (n = 3) poorly-formed somites, Myogenin expression was greatly reduced (Fig. 3E). The myogenic transcription factor Uncx4.1 is expressed on the posterior side of mature somites and in the PSM of wild type embryos at E9.5. In Notch1lbd/lbdembryos, expression in somites and PSM was lost (Fig. 3F). However, expression of Uncx4.1 was induced in brain in the absence of Notch1 signaling, as observed previously in embryos defective in signaling through all four Notch receptors [16]. We previously showed that cyclin D1 expression is markedly reduced in Notch1lbd/lbdembryos [19]. Importantly therefore, the Notch1 signaling defects observed in Notch1lbd/lbdembryos are not rescued by non-canonical Notch1 ligands that might bind to the large portion of the Notch1 extracellular domain that remains in Notch1lbd.
Maternal and zygotic Notchllbd/lbdmutant blastocysts implant and develop through gastrulation
Following oocyte-specific deletion of Pofut1 or RBP-Jκ null oocytes are fertilized and mutant blastocysts develop through gastrulation [20, 21]. Pofut1 [16] and RBP-Jκ [22] are essential for Notch signaling through all four Notch receptors. Pofut1 transfers fucose to Notch receptors and RBP-Jκ complexes with the cleaved ICD of all Notch receptors, but both activities might have effects that are independent of the Notch pathway. Thus it was of interest to determine if Notch1lbd/lbdblastocysts could develop and implant because Notch1 is expressed in oocytes, fertilized eggs and blastocysts [11, 12]. To obtain maternal and zygotic mutant blastocysts, females homozygous for the Notch1 floxed allele (Fig. 1A) and carrying a ZP3Cre transgene were generated (Fig. 4A). Notch1F/F:ZP3Cre females were mated with Notch1+/lbdor wild type males. Pups and E9.5 embryos were genotyped (Table 2). At birth, all pups from Notch1F/F:ZP3Cre by wild type crosses were heterozygous showing that the Cre recombinase was highly efficient since the Notch1Fallele was not transmitted. At E9.5, 29 embryos from 4 crosses included 15 mutants (Notch1lbd/lbd) and 14 heterozygotes (Notch1+/lbd). Therefore, eggs with Notch1 lacking the ligand binding domain were fertilized by sperm that also lacked functional Notch1 and gave the same number of E9.5 embryos as eggs fertilized with a Notch1+ sperm.
Notch1lbd/lbdmaternal and zygotic null embryos. (A) PCR of genomic DNA using primers 5F and 6R (Fig. 1A), and ZP3Cre primers. N1F: Notch1 floxed allele; Cre: ZP3Cre transgene. (B) – (C) E9.5 yolk sac with embryos from crosses between Notch1F/F:ZP3Cre females and Notch1+/lbdmales. (D) – (E) E8.75 Notch1+/lbdand Notch1lbd/lbdembryos. (F) – (G) E9.5 Notch1+/lbdand Notch1lbd/lbdembryos. Representative results from a total of 14 E8.75 embryos and 29 E9.5 embryos.
Mutant embryos derived from Notch1lbd/lbdeggs and therefore lacking maternal and zygotic transcripts of functional Notch1 were examined at E8.75 and E9.5. All Notch1lbd/lbdE9.5 embryos were surrounded by a yolk sac with defective vascularization (Figs. 4B, C). Notch1+/lbdand Notch1lbd/lbdembryos at E8.75 (8 vs. 6 from 2 litters) were morphologically indistinguishable (Figs. 4D, E). By E9.5, the Notch1lbd/lbdembryos were significantly smaller than controls, and the Notch1 null mutant phenotype was readily apparent (Figs. 4F, G). All developmental defects in Notch1lbd/lbdembryos arising from mutant blastocysts with or without maternal Notch1 transcripts appeared similar. In addition, mutant embryos were morphologically similar to controls at E8.75 (Fig. 4D, E). Therefore Notch1 signaling induced by canonical Notch ligands is not required for oogenesis, ovulation, fertilization or any of the developmental steps involved in blastogenesis, implantation or gastrulation.
Discussion
The Notch1lbdmutation is a novel Notch1 inactivating mutation that gives a null phenotype with a defective Notch1 that is expressed at normal levels at the cell surface. The Notch1 cleavage site mutant which is not efficiently cleaved by γ-secretase and is defective in Notch1 signaling [15] may also be expressed at the cell surface but this point has not been directly investigated. However, treatment of T cells with inhibitors of γ-secretase causes an accumulation of a membrane-bound Notch1 stable intermediate [23]. In Notch1lbd, removal of aa 290–481 from the Notch1 ECD does not inhibit Notch1 synthesis, trafficking or stability at the plasma membrane. Nevertheless, Notch1lbd exhibits reduced signaling through Delta1 and Jagged1 to the same extent as Notch1 null ES cells in a co-culture reporter assay. Moreover, the binding of Delta1 is reduced to the same extent as in cells that lack Notch1 altogether. This is consistent with in vitro binding experiments which identify EGF repeats 11 and 12 of Drosophila Notch as necessary for canonical ligand binding in vitro [3, 4]. However, the Drosophila Notch EGF11/12 deletion mutant has not been investigated for Notch signaling abilities, either in co-culture assays or,in vivo in the fly.
Notch1lbd/lbdembryos die at ~E10 with the same developmental phenotype as embryos that lack Notch1 [8, 9]. It is of interest that, despite the cell surface expression of most of the Notch1 extracellular domain in Notch1lbd, there appear to be no non-canonical Notch ligands that rescue mutant embryos at mid-gestation. Also of interest, is the fact that heterozygotes develop similarly to wild type mice, and thus Notch1lbd does not behave in a dominant negative fashion even though it is expressed at the cell surface in similar amounts to wild type Notch1. It will be of interest to see if Notch1+/lbdheterozygotes exhibit the more subtle Notch1 signaling defects observed in the inner ear [24], and in cell competition experiments [25] with other Notch1+/nullheterozygotes.
Whole mount in situ hybridization showed that Notch1lbd/lbdembryos exhibit markedly reduced expression of the Notch1 target genes Hes5 and cyclin D1 [19] and the somitogenic genes Myogenin and Uncx4.1. Similar results were observed in Notch1 null mutants [8, 9]. However, at E8.75 Notch1 transcripts were expressed at similar levels in mutant and wild type controls (Fig. 3A). Previous studies have shown that removal of Notch1 delays somitogenesis at the 3-5 somite stage around E8 showing that Notch1 signaling is active at this stage, even though there are no apparent changes in size, overall appearance, neurogenesis or cell death at E8 [9]. The embryos in Fig. 3A show that Notch1 gene expression in early embryogenesis is not solely controled by Notch1 signaling. This conclusion can also be drawn from the equivalent expression of Notch1 transcripts in Notch1lbd/lbdand Notch1+/+ ES cells (Fig. 1D). However, by E9.0 Notch1lbd/lbdembryos had markedly less Notch1 expression than controls (Fig. 2E). This may suggest that Notch1 signaling and Notch1 gene expression operate in a feedback loop at this stage as suggested from results of overexpression of Notch1 ICD in T cells [26, 27] or C2C12 cells [28]. However, overexpression experiments may induce abberrant regulation of the Notch1 gene and it is difficult to distinguish direct from indirect effects in vivo. Indirect effects on Notch1 gene expression are seen in Pofut1 [16] and Mib1 [17, 18] null embryos defective in global signaling that exhibit increased Notch1 gene expression in the PSM, the forming somite and the forebrain at ~E8.75, suggesting that Notch1 expression at that stage is negatively regulated via signaling through Notch2, Notch3 and/or Notch4. By E9.5, Notch1 gene expression is inhibited in the absence of Pofut1 [16] or Mind bomb 1 [17, 18]. Notch1 activation is also inhibited at E9.5 in embryos lacking RBP-Jκ [29].
The conditional Notch1 floxed allele allowed us to ask whether canonical Notch1 signaling is required for the generation of developmentally-competent eggs, or for fertilization, pre-implantation development, implantation or gastrulation. Previous experiments in which global Notch signaling was eliminated by the removal of Pofut1 or RBP-Jκ in oocytes suggest that Notch signaling is not required through any of the four mammalian Notch receptors until after gastrulation [20, 21]. However, Pofut1 transfers fucose to EGF repeats with a consensus sequence that is found in a number of proteins including Notch ligands and Cripto [30, 31]. While the presence of fucose on an EGF repeat is not required for the function of either Notch ligands [32] or Cripto [33], biological roles for O-fucose have only begun to be explored. In addition, recent experiments have shown that Notch lacking O-fucose may signal under certain circumstances [34, 48]. Similarly, Notch-independent functions of RBP-Jκ have been described [35, 36]. Finally, Notch1 is expressed in oocytes during oogenesis [37] and in ovulated eggs and developing blastocysts [11, 12], leading to the prediction that Notch1 signaling must be important for pre-implantation development [11, 12]. Thus it was important to examine this question directly. Our data clearly show that Notch1 signaling through canonical Notch ligands is in fact dispensable for oogenesis, ovulation, fertilization, blastogenesis, implantation and gastrulation (Fig. 4). They also show that expressing an inactive Notch receptor at the cell surface does not have an inhibitory effect on any of these developmental processes. However, non-canonical Notch1 signaling by a pathway yet to be discovered may be active in Notch1lbd/lbdoocytes or blastocysts. The fact that Notch1lbd is well-expressed at the cell surface but not responsive to canonical Notch ligands means it may be used to search for novel Notch1 signaling pathways that may be active in pre-implantation development.
Conclusion
In summary, we have shown that deletion of the mouse Notch1 ligand binding domain generates Notch1 of ~280 kDa that is well-expressed on the cell surface but cannot bind Delta1 nor be activated by Delta1 or Jagged1 in a co-culture signaling assay. Homozygous mutant embryos die at mid-gestation with defects similar to Notch1 null embryos. Oocyte-specific deletion of the ligand bindng domain does not impair oogenesis or development of maternal and zygotic embryos until after gastrulation.
Methods
Targeting of the Notch1 gene
To generate the Notch1 ligand binding domain deletion mutation Notch1lbd, an 1.6 kb region of genomic DNA containing exons 6 – 8 and two flanking sequences ~4.3 kb 5' (upstream) and ~2.9 kb 3' (downstream) were obtained by PCR from genomic DNA of WW6 ES cells [38] and cloned separately into pCR2.1 (T-vector, Invitrogen, Carlsbad, CA). A point mutation termed 12f was introduced into exon 8 by changing Thr466 to Ala [13]. The integrity of the three inserts was confirmed by DNA sequencing and they were subcloned between the three loxP sites in the pFlox vector [39] using BamHI, SalI and XhoI with HindIII, respectively (Fig. 1A). The targeting vector was linearized using PvuI (the PvuI site between XhoI and HindIII in pFlox had been removed during subcloning). After gel purification, the plasmid was electroporated into WW6 ES cells using a Bio-Rad Gene Pulser (Bio-Rad, Hercules, CA) at 400 V and 250 μF. Following selection with 250 μg/ml active G418 (Invitrogen, Carlsbad, CA), resistant colonies were screened for homologous recombination by PCR using Takara Ex Taq (Takara Mirus Bio, Madison, WI) and primers N1ES-gF: 5'-GCTTCCCGCCTCCACTGTGCTATTGATGTTTG-3' from upstream of the 5' insertion site and pFlx-382R: 5'-GTTCCTCTTGCTGAACCACACTGCTCGATATTG-3' from the pFlox vector, and confirmed using primers pFlx-3521F: 5'-CTGTGCCTTCTAGTTGCCAGCCATCTGTTG-3' from the pFlox vector and DM142: 5'-CTGAAGCCTTCTCGGCAGGTGCATACGTAG-3' from downstream of the 3' insertion. Two positive ES clones were further characterized by Southern blot analysis after digestion by HindIII or HindIII and EcoRI. The probe P1415 is a genomic DNA fragment obtained by PCR from exons 14 to 15 of Notch1 using primers N1-ex14F: 5'-GTACAAGTGACTGTGCCCCTGGGTG-3' and N1-ex15R: 5'- CTGTATATGGCAGAGGACAGTTGCACTTG-3' and was used to determine integration into the endogenous Notch1 locus. Targeted ES cells were microinjected into C57Bl/6 blastocysts to obtain chimeric mice. Chimeras were crossed with transgenic mice carrying Cre recombinase under the control of a weak CMV promoter termed MeuCre40 [14] to obtain heterozygous floxed Notch1 (Notch1+/F) mice. Oocyte-specific Notch1 deletion was obtained by crossing Notch1+/Fmice with transgenic mice carrying Cre recombinase under the control of the ZP3 promoter (ZP3Cre) as previously described [20, 39]. The deletion mutation was confirmed by Southern blot analysis after BamHI digestion using probe P1415 downstream of the 3' insertion site. Genotyping was performed from tail DNA or yolk sac DNA using forward primer 5F: 5'-GTATGTATATGGGACTTGTAGGCAG-3', and reverse primer 6R: 5'-CTATGAGGGGTCACAGGACCAT-3' that generate a 363 bp product from the wild type Notch1 allele and a 466 bp product from the floxed Notch1 allele, or primers 5F and 9R: 5'-CTTCATAACCTGTGGACGGGAG-3' that generate a 575 bp product from the Notch1 ligand binding domain deletion allele. ZP3Cre transgenic mice were genotyped as described [39].
Embryonic Stem Cell Isolation
Embryonic stem (ES) cell lines C1 (Notch1+/+), C2 (Notch1+/lbd) and A2 (Notch1lbd/lbd) were isolated from E3.5 blastocysts obtained from Notch+/lbdX Notch+/lbdcrosses as described [40] and genotyped by PCR from yolk sac DNA. Other ES cell lines used were WW6 [38] (Notch1+/+), and 290-2 Notch1 null cells [10] termed Notch1in32/in32 and kindly provided by Dr. G. D. Longmore. ES cells were routinely cultured on a STO SNL2 feeder cell layer [41] in ES medium (Knockout-DMEM supplemented with 15% fetal bovine serum; Gemini, West Sacramento, CA, 1 × non-essential amino acids, 1 × L-glutamine, 1000 U/ml ESGRO® (Chemicon, Temecula, CA), 50 μM ί-mercaptoethanol, 25 mM HEPES, penicillin (50 U/ml) and streptomycin (50 μg/ml). All reagents were ES-qualified and from SpecialtyMedia (Phillipsburg, NJ) except where mentioned). To remove feeder cells, ES cells were passaged on gelatinized plates for 2 – 3 generations at an 1:10 ratio. For growth curves, ES cells were plated on 24-well plates at 5 × 104 cells per well and incubated at 37°C in an incubater with 5% CO2. Cells from triplicate wells were trypsinized and counted after 24, 48, 72 and 96 h using a Z1 Coulter particle counter (Beckman Coulter, Fullerton, CA). Cells from each well were counted 3 times.
RT-PCR Analysis
Total RNA was isolated from ES cells using TRIZOL® (Invitrogen, Carlsbad, CA) followed by DNase I (Promega, Madison, WI) digestion according to the manufacturer's instructions. cDNA was prepared using the Takara RNA PCR Kit ver 3.0 (Takara Mirus Bio, Madison, WI). RT-PCR analysis was performed using Notch1 forward primer: 5'-GCCCTTTGAGTCTTCATACATCTG-3' and reverse primer: 5'-GACATTGGAACTCATTGATCTTGT-3'. PCR products (500 bp from Notch1lbdand 1076 bp from Notch1+) were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Gapdh was used as control (forward: 5'-ACCACAGTCCATGCCATCAC-3'; reverse: 5'-TCCACCACCCTGTTGCTGTA-3', product size: 452 bp).
Immunoblot Analysis
ES cells growing on gelatinized plates were washed with PBS and lysed in RIPA buffer (Upstate, Lake Placid, NY) containing the complete protease inhibitors 'cocktail' (Roche, Basel, Switzerland). After incubation for 30 min on ice, the lysate was microfuged and protein concentration of the supernate was determined by Bio-Rad D c protein assay (Bio-Rad, Hercules, CA). Lysates were resolved by 4–20% gradient SDS-PAGE, transferred to polyvinyldifluoride (PVDF) membranes and probed with Notch1 ECD antibody 8G10 (1:500; Upstate, Lake Placid, NY) to detect full length Notch1, or Notch1 antibody Val1744 (1:1000; Cell Signaling Technology, Beverly, MA) to detect activated Notch1. Horseradish peroxidase (HRP)-conjugated secondary antibodies were used to detect reactive bands visualized using Enhanced Chemiluminescence Reagent (Amersham Pharmacia Biotech, Piscataway, NJ). A β-Tubulin-III specific antibody (1:500; Sigma, St. Louis, MO) was used for loading control.
Flow Cytometry
For cell surface Notch1 detection, ES cells growing on gelatinized plates at 70–80% confluence were dissociated using phosphate buffered saline (PBS) -based enzyme-free dissociation solution (SpecialtyMedia, Lavellette, NJ) for 10 min at 37°C. After washing with 10 ml medium, 5 × 105 ES cells were incubated with 1 μg anti-Notch1 antibody (8G10, Upstate, Lake Placid, NY) in 100 μl FACS binding buffer (Hank's balanced salt solution (HBSS) containing 3% BSA, 0.05% sodium azide, and 1 mM Ca2+) for 1 h at 4°C in the dark, followed after washing with 1 ml FACS binding buffer, by incubation with 1:100 Alexa488-conjugated anti-hamster IgG antibody (Invitrogen, Carlsbad, CA) in 100 μl FACS binding buffer. After washing with 1 ml FACS binding buffer, the cells were suspended into 400 μl FACS binding buffer. Dead cells were excluded by staining with 7-AAD (BD Pharmingen, San Diego, CA). Flow cytometry was performed on a FACS Calibur flow cytometer (BD Biosciences, San Diego, CA). Data files were analyzed using Flowjo software (Tree Star, San Carlos, CA).
Notch Ligand Binding Assay
Soluble Notch ligand Delta1 with a human Fc tag (Delta1-Fc) [42, 43] was kindly provided by Dr. Gerry Weinmaster. HEK-293T cells expressing Delta1-Fc were cultured in αMEM (Invitrogen) containing 10% fetal bovine serum (Gemini). At 70–80% confluence, the medium was changed to 293 SFM II serum-free medium (Invitrogen) and culturing was continued. After 3 days, conditioned medium was collected, cellular debris removed by centrifugation, and the supernatant stored at 4°C. The concentration of ligand was determined by comparison with known concentrations of human IgG antibody (Jackson Immunoresearch, West Grove, PA) detected by chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ) after western blotting. For the binding assay, 70–80% confluent ES cells were dissociated from plates using PBS-based enzyme-free dissociation solution (SpecialtyMedia, Lavellette, NJ) for 10 min at 37°C. After washing with 10 ml medium, the single cell suspension of ES cells (5 × 105 cells) was incubated with 2 μg/ml Delta1-Fc in binding buffer (HBSS containing 3% BSA, 0.05% sodium azide, and 1 mM Ca2+) for 1 h at 4°C, followed after washing by incubation with 1:100 phycoerythrin (PE)-conjugated anti-human Fc antibody (Jackson Immunoresearch, West Grove, PA) for 30 min at 4°C. To inhibit Delta1 binding, 5 mM EDTA was added to the binding buffer. Flow cytometry was performed on a FACS Calibur flow cytometer. Ligand binding ability was determined by mean fluorescence intensity (MFI) of primary and secondary antibody binding minus the MFI of secondary antibody binding alone.
Co-culture Notch Signaling Assay
The co-culture Notch signaling assay was performed essentially as previously described [20]. In brief, duplicate cultures of ES cells (Notch1+/+,(C1) and Notch1lbd/lbd(A2)) on 6-well plates were co-transfected with the TP1-luciferase Notch reporter plasmid and a Renilla luciferase reporter (pRL-TK; Promega, Madison, WI) and empty vector using FuGENE 6 (Roche, Basel, Switzerland). At 16 h post-transfection, ES cells were overlaid with 1 × 106 Jagged1-expressing L cells or Delta1-expressing L cells or parental L cells. At ~40 h post-transfection, firefly and Renilla luciferase activities were quantitated in cell lysates using a dual luciferase assay kit (Promega, Madison, WI) on Autolumat Plus LB 953 (Berthold Technologies, Bad Wildbad, Germany) according to the manufacturer's instructions. Ligand-dependent Notch activation is expressed as fold-induction of normalized firefly luciferase activity obtained from Notch ligand versus L cell co-cultures. Co-culture assays with Notch1in32/in32 ES cells (290-2) compared to Notch1+/+ ES cells (C1) were performed by the same method in 12-well plates using Lipofectamine (Invitrogen) for transfection of plasmids.
Whole Mount in situHybridization
Embryos of E8.75, E9.0 or E9.5 from Notch1+/lbdcrosses were harvested and DNA was prepared from yolk sac for genotyping. Embryos for whole mount in situ hybridization were fixed in 4% formaldehyde in PBS overnight at 4°C. Whole-mount in situ hybridization was performed as previously described [16]. The hybridization probes used were: Notch1 full length, ~9.5 kb or ~4.7 kb [44]; Uncx4.1 ~1.7 kb [45], Hes5 ~1.3 kb [46]; and Myogenin ~1.5 kb [47]. Stained embryos were photographed in PBS through a phototube on Leica Wild M3Z stereomicroscope (Leica-Microsystems, Heerburgg, Switzerland) using a Canon S40 digital camera (Canon USA Inc., Lake Success, NY).
Abbreviations
- lbd:
-
ligand binding domain
- Pofut1:
-
protein O-fucosyltransferase 1
- ES:
-
embryonic stem
- EGF:
-
epidermal growth factor-like
- N1:
-
Notch1.
References
Lai EC: Notch signaling: control of cell communication and cell fate. Development (Cambridge, England). 2004, 131 (5): 965-973.
Schweisguth F: Regulation of notch signaling activity. Curr Biol. 2004, 14 (3): R129-38.
Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S: Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991, 67 (4): 687-699. 10.1016/0092-8674(91)90064-6.
Xu A, Lei L, Irvine KD: Regions of Drosophila Notch That Contribute to Ligand Binding and the Modulatory Influence of Fringe. 2005, 280 (34): 30158-30165.
Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P: The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002, 11 (21): 2615-2624. 10.1093/hmg/11.21.2615.
Mumm JS, Kopan R: Notch signaling: from the outside in. Dev Biol. 2000, 228 (2): 151-165. 10.1006/dbio.2000.9960. 2000/12/09
Fortini ME: Notch and presenilin: a proteolytic mechanism emerges. Curr Opin Cell Biol. 2001, 13 (5): 627-634. 10.1016/S0955-0674(00)00261-1. 2001/09/07
Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T: Notch1 is essential for postimplantation development in mice. Genes & development. 1994, 8 (6): 707-719. 10.1101/gad.8.6.707.
Conlon RA, Reaume AG, Rossant J: Notch1 is required for the coordinate segmentation of somites. Development (Cambridge, England). 1995, 121 (5): 1533-1545.
Hadland BK, Huppert SS, Kanungo J, Xue Y, Jiang R, Gridley T, Conlon RA, Cheng AM, Kopan R, Longmore GD: A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 2004, 104 (10): 3097-3105. 10.1182/blood-2004-03-1224.
Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M: A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004, 6 (1): 133-144. 10.1016/S1534-5807(03)00404-0.
Cormier S, Vandormael-Pournin S, Babinet C, Cohen-Tannoudji M: Developmental expression of the Notch signaling pathway genes during mouse preimplantation development. Gene Expr Patterns. 2004, 4 (6): 713-717. 10.1016/j.modgep.2004.04.003.
Ge CS: The O-fucose glycan in the ligand binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci U S A. 2008,
Leneuve P, Colnot S, Hamard G, Francis F, Niwa-Kawakita M, Giovannini M, Holzenberger M: Cre-mediated germline mosaicism: a new transgenic mouse for the selective removal of residual markers from tri-lox conditional alleles. Nucleic acids research. 2003, 31 (5): e21-10.1093/nar/gng021.
Schroeter EH, Kisslinger JA, Kopan R: Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998, 393 (6683): 382-386. 10.1038/30756. 1998/06/10
Shi S, Stanley P: Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2003, 100 (9): 5234-5239. 10.1073/pnas.0831126100.
Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, Lee J, Chitnis AB, Kim CH, Kong YY: Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development (Cambridge, England). 2005, 132 (15): 3459-3470.
Barsi JC, Rajendra R, Wu JI, Artzt K: Mind bomb1 is a ubiquitin ligase essential for mouse embryonic development and Notch signaling. Mech Dev. 2005, 122 (10): 1106-1117. 10.1016/j.mod.2005.06.005.
Stahl M, Ge C, Shi S, Pestell RG, Stanley P: Notch1-induced transformation of RKE-1 cells requires up-regulation of cyclin D1. Cancer research. 2006, 66 (15): 7562-7570. 10.1158/0008-5472.CAN-06-0974.
Shi S, Stahl M, Lu L, Stanley P: Canonical Notch signaling is dispensable for early cell fate specifications in mammals. Molecular and cellular biology. 2005, 25 (21): 9503-9508. 10.1128/MCB.25.21.9503-9508.2005.
Souilhol C, Cormier S, Tanigaki K, Babinet C, Cohen-Tannoudji M: RBP-Jkappa-dependent notch signaling is dispensable for mouse early embryonic development. Molecular and cellular biology. 2006, 26 (13): 4769-4774. 10.1128/MCB.00319-06.
Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T: Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development (Cambridge, England). 1995, 121 (10): 3291-3301.
Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC: Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Molecular and cellular biology. 2003, 23 (2): 655-664. 10.1128/MCB.23.2.655-664.2003.
Zhang N, Martin GV, Kelley MW, Gridley T: A mutation in the Lunatic fringe gene suppresses the effects of a Jagged2 mutation on inner hair cell development in the cochlea. Curr Biol. 2000, 10 (11): 659-662. 10.1016/S0960-9822(00)00522-4.
Visan I, Tan JB, Yuan JS, Harper JA, Koch U, Guidos CJ: Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat Immunol. 2006, 7 (6): 634-643. 10.1038/ni1345.
Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ: Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000, 13 (1): 73-84. 10.1016/S1074-7613(00)00009-1.
Matesic LE, Haines DC, Copeland NG, Jenkins NA: Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum Mol Genet. 2006, 15 (24): 3485-3497. 10.1093/hmg/ddl425.
Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J: Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997, 17 (10): 6057-6067.
Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL: Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Dev Dyn. 2007, 236 (9): 2594-2614. 10.1002/dvdy.21246.
Haines N, Irvine KD: Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003, 4 (10): 786-797.
Rampal R, Luther KB, Haltiwanger RS: Notch signaling in normal and disease States: possible therapies related to glycosylation. Curr Mol Med. 2007, 7 (4): 427-445. 10.2174/156652407780831593. 2007/06/23
Okajima T, Irvine KD: Regulation of notch signaling by o-linked fucose. Cell. 2002, 111 (6): 893-904. 10.1016/S0092-8674(02)01114-5. 2003/01/16
Shi S, Ge C, Luo Y, Hou X, Haltiwanger RS, Stanley P: The threonine that carries fucose, but not fucose, is required for cripto to facilitate nodal signaling. J Biol Chem. 2007, 282 (28): 20133-20141. 10.1074/jbc.M702593200.
Okajima T, Reddy B, Matsuda T, Irvine KD: Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 2008, 6 (1): 1-10.1186/1741-7007-6-1.
Levy OA, Lah JJ, Levey AI: Notch signaling inhibits PC12 cell neurite outgrowth via RBP-J-dependent and -independent mechanisms. Dev Neurosci. 2002, 24 (1): 79-88. 10.1159/000064948.
Dumont E, Fuchs KP, Bommer G, Christoph B, Kremmer E, Kempkes B: Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene. 2000, 19 (4): 556-561. 10.1038/sj.onc.1203352.
Hahn KL, Johnson J, Beres BJ, Howard S, Wilson-Rawls J: Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development (Cambridge, England). 2005, 132 (4): 817-828.
Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor SM, Stanley P: WW6: an embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proceedings of the National Academy of Sciences of the United States of America. 1995, 92 (16): 7357-7361. 10.1073/pnas.92.16.7357. 1995/08/01
Shi S, Williams SA, Seppo A, Kurniawan H, Chen W, Ye Z, Marth JD, Stanley P: Inactivation of the Mgat1 gene in oocytes impairs oogenesis, but embryos lacking complex and hybrid N-glycans develop and implant. Molecular and cellular biology. 2004, 24 (22): 9920-9929. 10.1128/MCB.24.22.9920-9929.2004.
Roach ML, McNeish JD: Methods for the isolation and maintenance of murine embryonic stem cells. Methods Mol Biol. 2002, 185: 1-16. 2002/01/05
Ioffe E, Stanley P: Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proceedings of the National Academy of Sciences of the United States of America. 1994, 91 (2): 728-732. 10.1073/pnas.91.2.728. 1994/01/18
Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA: Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998, 21 (1): 63-75. 10.1016/S0896-6273(00)80515-2.
Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G: Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000, 2 (8): 515-520. 10.1038/35019553.
Nye JS, Kopan R, Axel R: An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development (Cambridge, England). 1994, 120 (9): 2421-2430. 1994/09/01
Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R: Dynamic expression and essential functions of Hes7 in somite segmentation. Genes & development. 2001, 15 (20): 2642-2647. 10.1101/gad.930601. 2001/10/20
Akazawa C, Sasai Y, Nakanishi S, Kageyama R: Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992, 267 (30): 21879-21885. 1992/10/25
Wright WE, Sassoon DA, Lin VK: Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989, 56 (4): 607-617. 10.1016/0092-8674(89)90583-7. 1989/02/24
Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P: Roles of pofut1 and o-fucose in Mammalian notch signaling. J Biol Chem. 2008, 283 (20): 13638-13651. 10.1074/jbc.M802027200.
Acknowledgements
The authors gratefully acknowledge the excellent technical assistance of Wen Dong. We also thank Dr. Shaolin Shi (Mt. Sinai School of Medicine, New York) for reagents, advice and comments on the manuscript, and we thank all those who supplied mice, cell lines and probes as noted in the text. This work was supported by NIH grant RO1 NCI 95022 to PS and partial support was provided by the Albert Einstein Cancer Center grant NCI PO1 13330.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CG and PS conceived the project and designed the experiments. CG performed the experiments in Figures 1, 2 and 4 and Tables 1 and 2 and wrote the paper. TL did the in situ hybridization experiments (Figure 3 and text) and helped to write the paper. XH did co-culture Notch signaling assays. All authors interpreted data and wrote the paper. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ge, C., Liu, T., Hou, X. et al. In vivo consequences of deleting EGF repeats 8–12 including the ligand binding domain of mouse Notch1. BMC Dev Biol 8, 48 (2008). https://doi.org/10.1186/1471-213X-8-48
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-213X-8-48