Abstract
Background
Myf5 is one member of the basic helix-loop-helix family of transcription factors, and it functions as a myogenic factor that is important for the specification and differentiation of muscle cells. The expression of myf5 is somite- and stage-dependent during embryogenesis through a delicate regulation. However, this complex regulatory mechanism of myf5 is not clearly understood.
Results
We isolated a 156-kb bacterial artificial chromosome clone that includes an upstream 80-kb region and a downstream 70-kb region of zebrafish myf5 and generated a transgenic line carrying this 156-kb segment fused to a green fluorescent protein (GFP) reporter gene. We find strong GFP expression in the most rostral somite and in the presomitic mesoderm during segmentation stages, similar to endogenous myf5 expression. Later, the GFP signals persist in caudal somites near the tail bud but are down-regulated in the older, rostral somites. During the pharyngula period, we detect GFP signals in pectoral fin buds, dorsal rostral myotomes, hypaxial myotomes, and inferior oblique and superior oblique muscles, a pattern that also corresponds well with endogenous myf5 transcripts. To characterize the specific upstream cis-elements that regulate this complex and dynamic expression pattern, we also generated several transgenic lines that harbor various lengths within the upstream 80-kb segment. We find that (1) the -80 kb/-9977 segment contains a fin and cranial muscle element and a notochord repressor; (2) the -9977/-6213 segment contains a strong repressive element that does not include the notochord-specific repressor; (3) the -6212/-2938 segment contains tissue-specific elements for bone and spinal cord; (4) the -2937/-291 segment contains an eye enhancer, and the -2937/-2457 segment is required for notochord and myocyte expression; and (5) the -290/-1 segment is responsible for basal transcription in somites and the presomitic mesoderm.
Conclusion
We suggest that the cell lineage-specific expression of myf5 is delicately orchestrated by multiple modules within the distal upstream region. This study provides an insight to understand the molecular control of myf5 and myogenesis in the zebrafish.
Similar content being viewed by others
Background
Members of the basic helix-loop-helix (bHLH) family of transcription factors, such as Myf5, MyoD, Myogenin, and MRF4, are crucially important in the specification and differentiation of skeletal muscle progenitors [1]. These myogenic regulatory factors (MRFs) activate muscle-specific transcription by binding to an E-box in the promoter of numerous muscle-specific genes [2, 3]. MRF genes are expressed in zebrafish somites in a characteristic temporal sequence, with myf5 at 7.5 hours postfertilization (hpf) [4], myod at 8 hpf [5], and myogenin at 10.5 hpf [5]. The same temporal sequence occurs in mice [1]. These observations indicate that myf5 is the first MRF expressed during vertebrate myogenesis.
Mechanisms that lead to Myf5 activation at multiple sites in mouse embryos have been described [6, 7]. Yeast artificial chromosomes (YAC) [6] and bacterial artificial chromosomes (BAC) [7] have been used to map the promoter of mouse myf5, suggesting that several different cis-regulatory elements are required to activate myf5 expression in different cells at different developmental times. An enhancer at -6.6 kb is required for myf5 expression in the epaxial domain [8]. A 270-bp core enhancer at -57 kb directs myf5 expression in limbs and maintains myf5 expression in somites [9]. In Xenopus, two negative regulatory elements have been identified: an interferon regulatory factor-like DNA binding element that down-regulates Xmyf5 expression in differentiating myocytes [10], and a distal TCF-3 binding site by which Wnt/β-catenin signaling restricts Xmyf5 expression to the midline mesoderm [11]. A T-box binding site mediates dorsal activation of Xmyf5 transcription and is involved in the regulation of muscle development [12]. Using transient expression of transgenes, we previously identified some cis-elements that regulate zebrafish myf5 [4, 13, 14]. Recently, Lee et al. [15] demonstrated that Foxd3 binds to the -82/-62 regulatory module and regulates zebrafish myf5 expression during early somitogenesis. These observations highlight the complicated and dispersed nature of the upstream elements that control somite- and stage-specific expression of myf5.
To elucidate the nature of this finely tuned control mechanism, we needed a transgenic line that recapitulates the specific endogenous expression pattern of myf5. Such a line requires a transgene that contains a very long upstream region of myf5. We modified the techniques used in mice [16, 17] and present here a highly efficient method for engineering zebrafish BAC. The BAC is an Escherichia coli F factor-based vector that is capable of propagating cloned DNA fragments up to 300 kb long [18]. Previously, Jessen et al. [19] reported a homologous recombination technique for BAC cloning to generate transgenic zebrafish. This technique, however, is rather laborious because it requires a chi-based plasmid with a very large recombination targeting region. With our new method, we efficiently generate transgenic lines containing a 156-kb genomic sequence of myf5 (80-kb upstream and 70-kb downstream segments) replaced with green fluorescent protein (GFP) in the coding region. We find that the 156-kb genomic sequence is long enough to recapitulate the endogenous myf5 transcription pattern during the somitogenesis and pharyngula development stages. To characterize the functions of individual cis-regulatory elements, we also generated several transgenic lines that carry various lengths of the zebrafish myf5 upstream sequence. Comparing the GFP expression patterns of these lines, we identified and characterized the functions of upstream regulatory elements, including a repressive element and tissue-specific enhancers for jaw and fin muscles, bones, eyes, somites, olfactory organs, and the presomitic mesoderm. Whole-mount in situ hybridization reveals that endogenous zebrafish myf5 transcripts are first detectable in the presomitic mesoderm [4]. In contrast, myf5 expression has not been observed by in situ hybridization in the presomitic mesoderm of mouse embryos, although weak signals have been detected by reverse transcriptase-polymerase chain reaction (RT-PCR) [20, 21]. These comparisons illustrate the advantage of our transgenic lines for studying the initiation and regulation of myf5, particularly in the presomitic mesoderm.
Results
Genomic organization of zebrafish myf5is conserved with mammals
We cloned the myf5 genomic locus using a sequential screening method. We screened 10 primary pools of zebrafish BAC library clones using PCR; one (P1) was positive. We then screened 48 secondary pools derived from P1 and found 5 positive pools. Subsequent screening of the positive pools identified a single myf5-positive BAC clone with a 156-kb insert as indicated by PFGE. We searched sequence databases from the Sanger Institute and identified a contig, ctg9418, which contains the entire sequence of the myf5 BAC clone. BLAST analysis using the myf5 coding region and junction sequences flanking the T7 and SP6 sites showed that the BAC clone spans the sequences of ctg9418 from 1460 to 1304 kb, with the coding sequence starting at 1380. Thus, the myf5 5' and 3' regions contained in this BAC clone extend approximately 80 kb and 70 kb, respectively, beyond the coding sequence.
Radiation hybrid mapping of zebrafish myf5 revealed that myf5 is located on linkage group 4 (LG4), between expressed sequence tags (ESTs) fb62d08 and fb78c03, and at 5.87 CR from EST marker z9667 (data not shown). This syntenic relationship indicates that the region of zebrafish LG4 between EST markers fa05f06 and fk68a09 (including myf5) is conserved with human chromosome 12q13–12q21.
DNA fragments of myf5:GFPare inherited
We generated several transgenic lines by microinjecting zebrafish embryos with myf5:GFP BAC segment and raising them to adulthood. We identified germ-line transmission by looking for GFP fluorescence in embryos from crossing with wild-type fish. We generated 4 lines with the entire myf5 upstream segment (the 156-kb group: 80k-5, -18, -21, and -23; Fig. 1A), 4 lines with -9977/-1 (10k-2, -13, -9R, -15R; Fig. 1B), 4 lines with -6212/-1 (6k-9R, -10R, -11R, -16R; Fig. 1B), 3 lines with -2937/-1 (3k-18R, -92R, -104R; Fig. 1B), 3 lines with -2456/-1 (2.4k-3, -8, -55; Fig. 1B), and 2 lines with -290/-1 group (0.3k-14R, -112R; Fig. 1B). The F2 segregation frequencies for these transgenic lines ranged from 47.5 to 52.9%, indicating a single insertion site of transgene for each line.
Construction of a myf5:GFP bacterial artificial chromosome (BAC) and deletion constructs for germ-line transmission in zebrafish. (A) Strategy for constructing a myf5 BAC clone containing the green fluorescent protein (GFP) reporter. (Top) The genomic organization of the myf5 that contains 3 exons (E1, E2, and E3) and 2 introns (I1 and I2). (Bottom) The resulting p(myf5(80K):GFP) clone contains the myf5 upstream 80-kb regions fused with the GFP reporter gene. The primers ZMFP-117F, GFP-R, Kan-F, and ZMF-1000R were used to check recombinants. (B) Deletion constructs used in this study. Plasmid pZMYP-2456E was described by Wang et al. [13]. Thick lines and crossed boxes represented plasmid vectors and myf5 promoters, respectively. Numbers above the boxes indicate the nucleotide positions relative to the transcription start site of zebrafish myf5. GFP, green fluorescent protein; ITR, inverted terminal repeats of adeno-associated virus; SVpA, polyadenylation signal of SV40.
The 156-kb genomic sequence of myf5drives GFP expression in muscle precursors
In embryos derived from myf5:GFP transgenic lines with the 156-kb genomic sequence (an upstream 80-kb segment and a downstream 70-kb segment; Fig. 1A), GFP fluorescence first appears very weakly at 7.5 hpf (data not shown), reaches detectable levels in the segmental plate by 10.5 hpf (Fig. 2A), and expands to 14 somite pairs in 16 hpf embryos (Fig. 2B). At 16 hpf, the GFP signals are strong and mainly restricted to the somites and segmental plates and fluorescence is reduced in the more rostral (older) somites. Prominent GFP signals also appear in the adaxial cells (Fig. 2B). The GFP mRNA in embryos derived from myf5:GFP transgenic lines were also detected by using digoxigenin (DIG)-labeling GFP riboprobes. The GFP transcripts were detectable at 7.5 hpf (Fig. 2F) and 10.5 hpf (Fig. 2G), which matches the spatial and temporal pattern of endogenous myf5 expression as indicated by mRNA in situ hybridization (Figs. 2C–2E) [4]. These results indicate that expression of GFP in the myf5:GFP transgenic line recapitulates endogenous myf5 expression.
Green fluorescent protein (GFP) expression in Tg ( myf 5(80K):GFP) transgenic embryos recapitulates endogenous myf5 expression in muscle precursors. GFP fluorescence is detected in the presomitic mesoderm of embryos by 10.5 hours postfertilization (hpf) (A), in the somites and the presomitic mesoderm in 16 hpf embryos (B). Endogenous myf5 transcripts (C,D,E) and GFP mRNA (F,G) were detectable at 7.5, 10.5, and 16 hpf. (A,D,G) Dorsal views, rostral to the left; (B,C,E,F) side views, rostral to the left, dorsal to the top. Scale bars: 100 μm in all panels.
By 28 hpf, GFP fluorescence is absent from rostral (older) somites (Figs. 3A and 3B). Cross-sections reveal that GFP-positive cells are distributed throughout the myotome (Fig. 3C). We use the F59 antibody to label slow muscle fibers and find that both slow and fast muscle fibers express GFP in the myf5:GFP transgenic line (Figs. 3D and 3E). In addition, we find no significant differences when comparing the GFP expression patterns of the four independent myf5:GFP lines (80k-5, -18, -21, and -23), indicating that expression of the transgene is unaffected by its chromosomal location.
Tg ( myf 5(80k):GFP) transgenic embryos express green fluorescent protein (GFP) in both slow and fast muscle fibers. (A,B) GFP expression in somites labeled with the F59 antibody. White dash lines indicate the location of head. (B) Higher magnification view of the boxed region shown in A. (C-E) Cross-section along the plane indicated by the white line in panel (B). GFP signals are observed in both fast (C, green signals) and slow muscle fibers (F, yellow signals). (A,B) Side views, rostral to the left, dorsal to the top; (C-D) dorsal to the top. 28 hpf. Scale bars: 400 μm in A; 200 μm in B; 100 μm in C-E.
GFP expression reveals the development of head skeletal muscles
By the end of segmentation stages, pectoral fin (pm) and myotomal (m), dorsal rostral myotomal (drm), muscle precursors express GFP (Figs. 4A and 4B). By 36 hpf, in addition to muscle precursors of pm, m, and drm, two eye muscle precursors, the superior oblique (so) and inferior oblique (io), also express GFP (Figs. 4C–4E) and endogenous myf5 transcripts (Figs. 4F–4H). At 60 hpf, GFP signals are observed in almost all cranial muscle precursors, including the adductor hyomandibulae (ah), adductor mandibulae (am), adductor operculi (ao), constrictor hyoideus ventralis (chv), dilatator operculi (do), inferior oblique (io), lateral rectus (lr), medial rectus (mr), sternohyoideus (sh), superior oblique (so), and transverse ventralis (tv1–5) (Figs. 5A and 5B). Compared with the broad expression of GFP fluorescence, expression in both GFP and endogenous myf5 transcripts is restricted to a tiny spot on each pharyngeal arch at this stage on lateral views (Figs. 5C and 5F, arrows) by using whole-mount in situ hybridization. However, either GFP or myf5 signals were observed at this stage on ventral views (Figs. 5D and 5G). Thus, we conclude that this inconsistency between GFP mRNA and fluorescence should be due to the stability of GFP that persists even though transcription of myf5 has ended. Based on these observations, we propose that most and possibly all cranial muscle precursors transiently express myf5 before 60 hpf, although myod expression is still evident at this stage (Fig. 5E).
Expression of green fluorescent protein (GFP) in Tg ( myf 5(80K):GFP) transgenics matches the dynamic pattern of endogenous myf5 expression in cranial muscles. (A-D) GFP fluorescence is apparent in pectoral fin muscle (pm), dorsal rostral muscle (drm), and hypaxial muscle (hy). (C-E) GFP fluorescence is detected in the occipital somite (os; precursors of sternohyoideus, sh) and some cranial muscles, such as the superior oblique (so) and inferior oblique (io). (F-H) Endogenous myf5 transcripts are also detected in cranial muscles, including so and io by whole-mount mRNA in situ hybridization. (A,C,F) Side views, rostral to the left, dorsal to the top; (B,D,G) dorsal views, rostral to the left; (E,H) ventral views, rostral to the left. Scale bars: 200 μm.
Green fluorescent protein (GFP) persists in cranial muscles of Tg ( myf 5(80k):GFP) transgenics. (A,B) GFP fluorescence is apparent in adductor hyomandibulae (ah), adductor mandibulae (am), adductor operculi (ao), constrictor hyoideus ventralis (chv), dilatator operculi (do), inferior oblique (io), lateral rectus (lr), medial rectus (mr), sternohyoideus (sh), superior oblique (so), and transverse ventralis (tv1–5) at 60 hours postfertilization (hpf). (C, D) Endogenous myf5 transcripts and (F, G) GFP mRNA are restricted to four spots by 60 hpf using whole-mount mRNA in situ hybridization. (E) At 60 hpf, myod transcripts are detected in most if not all cranial muscles. (A,C,E,F) Side views, rostral to the left, dorsal to the top; (B,D,G) ventral view, rostral to the left. Scale bars: 200 μm in all panels.
Proximal element regulates myf5expression in the presomitic mesoderm
A unique characteristic of myf5 expression, compared with other known MRFs, is its strong and widespread expression in the presomitic mesoderm (Fig. 2B). We used the transgenic lines to identify the regulatory sequences required for this aspect of myf5 expression. At 16 hpf, Tg(myf5(80K):GFP) embryos show strong GFP expression in adaxial cells, presomitic mesoderm, and developing somites (Fig. 6A). Transgenic lines carrying shorter upstream segments, -9977/-1 Tg(myf5(10K):GFP) (Fig. 6B), -6212/-1 Tg(myf5(6K):GFP), -2937/-1 Tg(myf5(3K):GFP), -2456/-1 Tg(myf5(2.4K):GFP) (Table 1), also show strong GFP signals in the presomitic mesoderm. However, in the transgenic line carrying the -290/-1 segment Tg(myf5(0.3K):GFP), the GFP signal in the presomitic mesoderm is weak (Table 1). These results suggest that the minimal enhancer that regulates myf5 expression in the presomitic mesoderm is located within the -290/-1 segment.
A proximal element regulates myf5 expression in the presomitic mesoderm. Green fluorescent protein (GFP) fluorescence is detected in the presomitic and somitic mesoderm of transgenic embryos harboring (A) -80 kb, Tg(myf5(80K):GFP) or (B) -10 kb, Tg(myf5(10K):GFP). Tg(myf5(10K):GFP) embryos also express GFP in the notochord. Dorsal views, rostral to the left. Scale bars: 200 μm.
Repressive element located at -80/-10 kb blocks GFP expression in notochord driven by module -2937/-2457
We observe no GFP signal in the notochords of embryos derived from line Tg(myf5(80K):GFP). However, Tg(myf5(10K):GFP) (Fig. 7A), Tg(myf5(6K):GFP) (Fig. 7B), and Tg(myf5(3K):GFP) (Figs. 7C and 7D) embryos have GFP expression in their notochords, suggesting that an element located within -80/-10 kb normally represses myf5 expression in notochord. We also observe no GFP in notochords of embryos derived from lines Tg(myf5(2.4K):GFP) and Tg(myf5(0.3K):GFP) (Table 1), suggesting that the -2937/-2457 segment contains a notochord enhancer element. To test this hypothesis, we constructed and microinjected three different GFP expression plasmids: pEGFPmTATA contains a TATA-box of CMV fused to EGFP, pEGFPm(2937/2457) contains one copy of segment -2937/-2457, and pEGFPm(2457/2937) contains segment -2937/-2457 in the opposite orientation (Additional file 1). Only 5.1% (6 of 104) of pEGFPmTATA-injected embryos are GFP-positive, and in no cases is GFP expressed either in notochord or myocytes (Additional file 1). However, 48.4% of pEGFPm(2937/2457)-injected embryos and 44.5% of pEGFPm(2457/2937)-injected embryos express GFP in both notochord and myocytes (Additional file 1). Thus, the -2937/-2457 segment is a typical, orientation-independent enhancer module for myf5 expression in notochord and myocytes.
The myf5 upstream region contains modules that repress expression in notochord. Green fluorescent protein (GFP) fluorescence is detected in the notochord of transgenic embryos harboring (A) -10 kb, Tg(myf5(10K):GFP), (B) -6 kb, Tg(myf5(6K):GFP), or (C-D) -3 kb, Tg(myf5(3K):GFP). Side views, rostral to the left. Scale bars: 400 μm in A; 200 μm in B, C, D.
Multiple modules regulate myf5expression in the spinal cord, bones, eyes, and olfactory pits
In embryos derived from Tg(myf5(6K):GFP), GFP fluorescence appears in adductor mandibulae (am) and dorsally caudal to the hindbrain at 48 hpf (Fig. 8A). By 72 hpf, GFP fluorescence is stronger and extends farther caudally (Fig. 8B), including cells of both the surface ectoderm and spinal cord (Figs. 8C and 8D). From 21–60 hpf, the embryos derived from Tg(myf5(6K):GFP) show GFP fluorescence in bone, including the basihyal, sternohyal, and palatoquadrate bones, and Meckel's cartilage (Figs. 8E–8G), and in the eyes (Fig. 8H). We find no significant differences when comparing the GFP expression patterns of the four independent myf5:GFP lines (6k-9R, -10R, -11R, -16R), indicating that expression of the transgene is unaffected by its chromosomal location. In the head of embryos derived from Tg(myf5(3K):GFP), GFP is expressed primarily in the eyes (Fig. 8I) and olfactory placode (Fig. 8J), suggesting that eye and olfactory enhancers are located within the -2937/-291 segment. Again, we believe that expression of the transgene is unaffected by its chromosomal location, because no significant differences were observed when comparing the GFP expression patterns of the three independent myf5:GFP lines (3k-18R, -92R, -104R).
The myf5 upstream region contains modules that regulate expression in spinal cord, bones, eyes and olfactory-pits. (A,B) Green fluorescent protein (GFP) fluorescence at 48 hours postfertilization (hpf) (A) and 72 hpf (B); the star in A indicates the location of hindbrain. (C,D) Cross-sections along the plane indicated by lines C and D in B, GFP signals are apparent in spinal cord (sc) and surface ectoderm. (E-J) GFP expression is observed in bones at 21 dpf (E and F), in bones at 60 dpf (G and H) in eyes (H and I), and olfactory pits (J). (K) The same embryo as (J) with brightfield illumination. am, adductor mandibulae; bh, basihyal; mc, Meckel's cartilage; n, notochord; op, olfactory pits; pq, palatoquadrate; sc, spinal cord. (A,B,E,H) Lateral views, rostral to the left, dorsal to the top; (F,G) ventral views, rostral to the left; (I-K) frontal views, dorsal to the top. Scare bars: 500 μm in A, E-H; 250 μm in I; 200 μm in B; 100 μm in C, D, J, K.
Discussion
Stable transgenic lines provide greater sensitivity for studies of gene regulation and confirm transient transgenesis studies
To understand the complex spatial and temporal regulation of myf5 expression, we analyzed the function of the zebrafish myf5 promoter using transgenic constructs. Our previous analysis of embryos injected with various lengths of myf5 promoter driven reporter genes (transient transgenesis) showed that -9977/-1 [22], -6212/-1 [4], and -2937/-1 [13] produce GFP-positive signals in the notochord, whereas no fluorescence is observed in the notochord when embryos are injected with -2456/-1 [13]. These observations suggest that the notochord-specific element is located in the -2937/-2457 segment, consistent with the germ-line transmission analysis of our present study (Fig. 9).
The regulatory elements for muscle lineage-specific expression, including slow and fast myotomes, are controlled by Hedgehog signaling and myocyte enhancer factor 2 (MEF2) [8, 23–25]. We propose that the somite- and myotomal-restricted enhancer element is located within the -290/-1 segment (Fig. 9). Thus, it is important to look for the putative myotomal enhancers, such as Hedgehog-responsive elements and MEF2-binding sites within this segment. After sequence analysis, we find that there are one MEF2-binding (-277/-268) and two Gli-binding (-252/-243 and -196/-183) sites located within the -290/-1 segment. In addition, our previous studies showed that the -82/-62 cassette that contains a binding site for the cognate trans-acting factor Foxd3 [15] is able to drive transient expression in somites [14]. On the basis of these observations, we propose that -82/-62 motif is a key element for driving somite specificity, and multiple elements within the -290/-1 segment such as MEF2- and Gli-binding sites are responsible for myotomal expression. Together, these results demonstrate that transient analysis is suitable for rapid identification of putative cis-elements, whereas germ-line transmission studies provide great sensitivity and enable confirmation.
Many myf5regulatory modules are similar between mouse and zebrafish
Few reports have proposed using transgenic animals to study the delicate transcriptional regulation of myf5. Only three species (mice, zebrafish, and Xenopus) have been documented so far. With limited information on the interactions between trans-acting factors and their binding sequences upstream of the myf5 gene, the alternative ways to study transcriptional mechanistic conservation are those making comparisons of sequence similarities and genomic organizations of the myf5 gene among these three known species. As in zebrafish, myf5 expression is regulated by multiple upstream sites in mouse embryos [7, 26]. Multiple enhancers are distributed throughout a 90-kb region of the mouse mrf4/myf5 locus, including a limb enhancer at -58/-48 kb, two head muscle enhancers at -88/-63 and -45/-23 kb, a repressive element at -58/-8.8 kb, a central nervous system-specific enhancer at -0.5/-0.1 kb, a hypaxial myotome enhancer at 0.5/3.5 kb, an epaxial myotome enhancer at -5.6/-4.6 kb, and branchial enhancers at -1.5/-0.5 and 0.5/3.5 kb [6, 7, 27]. Gene regulation of Myf5 in frog is apparently quite different, because only a relatively short region of flanking DNA (about 1.2 kb) is sufficient to drive endogenous Xenopus Xmyf5 expression at gastrula stages. Two negative regulatory elements (an interferon-like regulatory factor and a distal TCF-3 binding site) have been identified in the Xmyf5 promoter [10, 11]. It seems likely that each species (i.e., mice, zebrafish and Xenopus) uses distinct mechanisms, given the developmental differences in timing and signaling.
The genomic organizations of myf5 regulatory modules are also conserved between mice and zebrafish. As in mouse, the zebrafish regulatory modules that are responsible for lineage-specific expression of myf5 are distributed throughout a large (80-kb) region of the mrf4/myf5 locus (Fig. 9). The locations and functions of some enhancers are similar to those of mouse myf5. For example, the fin muscle enhancer and the cranial muscle enhancer at -80/-10 kb probably correspond to the mouse limb and head muscle enhancers at -88/-23 kb, a repressive one at -10/-6 kb is similar to the mouse repressor at -58/-8.8 kb, and a spinal cord enhancer at -6/-3 kb is similar to the mouse central nervous system-specific enhancer at -0.5/-0.1 kb (Fig. 9).
On the other hand, however, we also identified enhancers that have not been described in mouse, including a bone enhancer at -6/-3 kb, a notochord enhancer at -3/-2.4 kb, and a presomitic mesoderm enhancer at -290/-1 bp (Fig. 9). Epaxial- and hypaxial-specific enhancers have been identified in mouse myf5 [6, 7, 27], but we have not seen muscle lineage-specific enhancers, especially slow and fast muscle enhancers in zebrafish, with the exception of limb and head muscle enhancers. This finding suggests that more transgenic lines that carry shorter elements should be generated, especially deletions within the -80/-10 kb and -10/-6 kb regions. Taken together, we propose that the regulation of zebrafish myf5 is more similar to mouse myf5 than that of Xenopus.
Excellent experimental materials to study transcriptional regulation on myf5
Myf5 is the first member of the MRF family expressed during somitogenesis in zebrafish. Knockdown of myf5 in zebrafish results in malformation of somites and brain defects, indicating that trunk and head myogenesis is impaired, underscoring the importance of understanding the regulation of myf5. Numerous factors have been implicated as upstream regulators of myf5, including extracellular signals, such as shh, wingless (wnt), and fibroblast growth factor (FGF) [28, 29]. Shh, produced by the notochord and floor plate of the neural tube, and Wnt proteins, produced by the dorsal neural tube and surface ectoderm, have been implicated in the maintenance of mouse myf5 [8, 21, 30, 31], Xenopus Xmyf5 [32], and zebrafish myf5 [33] expression. Together with our previous identification of the cognate trans-acting factor, Foxd3 [15], these results demonstrate that the complex spatial and temporal pattern of myf5 gene expression is regulated by multiple upstream regulatory modules.
Conclusion
We generated several transgenic lines of zebrafish that contain various lengths of zebrafish myf5 upstream genomic sequence linked to the GFP reporter gene. We demonstrate that the 156-kb genomic sequence (an upstream 80-kb and a downstream 70-kb segment) is able to recapitulate the pattern of endogenous myf5 expression. By dissecting this upstream region, we further show that tissue-specific regulatory elements are organized as modules in various regions of the 5'-flanking sequence. These transgenic lines not only provide excellent materials for studying the regulatory mechanism of myf5, but they also will facilitate mutant screens to identify novel genes that regulate somitogenesis and more detailed studies to the morphogenesis of somites, presomitic mesoderm, and cranial muscles.
Methods
Animals
Embryos were produced using standard procedures [34] and were staged according to standard criteria [35] or by hpf at 28°C. The wild-type line used in this study was AB. Line and gene names follow the zebrafish nomenclature conventions [36].
BAC library screening
The zebrafish BAC library was obtained from the RZPD [37], and the screening protocols followed the manufacturer's instructions, with minor modifications. The primary BAC library pools were screened by PCR using the zebrafish myf5 intron 1-specific primers 1261F (5'-TGTTCATTCACTCATTTTCTTTTCA-3') and 2582R (5'-GCAGTCTTCCTACAATGACAA-3'). The positive clones isolated from the primary pools were further confirmed by screening the secondary pools to isolate a myf5-containing BAC clone.
Pulsed-field gel electrophoresis
DNA from a myf5-containing BAC clone was extracted; digested with EcoRI, HindIII, and SacI; and analyzed by 0.8% Agarose pulsed-field gel electrophoresis (PFGE; Biometra). The electrophoresis conditions were 200 volts at 10°C for 24 h with electrode angles at 120°, and rotor speed of 2–6 s. After electrophoresis, the BAC DNA size was analyzed with Kodak 1D image software.
Bioinformatics
We used the zebrafish myf5-specific primers 1261F and 2582R for mapping myf5 against the LN54 radiation hybrid (RH) panel. The RH panel was scored according to Hudson et al. [38] using the public web site [39]. For screening the myf5-containing BAC clone, the junctions of BAC DNA were sequenced using T7 and SP6 primers. These junction sequences and adjacent coding regions were analyzed by BlastN [40], and the location of the myf5-containing BAC clone was characterized and named myf5(80K).
Generation of a myf5(80K) clone containing the GFP reporter
Plasmid p(myf5(80K):GFP) contains the approximately 80-kb region around myf5 fused to GFP (Fig. 1A). Basically, we followed the protocols described by Lee et al. [17], with some modifications. The cassette used for targeting the myf5 locus was amplified from template pZMYP-82E [4] with primers ZMFP-82F (5'-CTCTTAGCTCTGTCCTGGCCA-3') and Kan-817R (5'-ATTTACAAATGAGCAAGCAGTGTGAATAAAGCGTTGGCCTGAGTCGGTCATTTCGAACCCCAG-3') by using Deep Vent (NEB) to carry out PCR at the following conditions: 94°C, 40 s; 58°C, 30 s; 72°C, 150 s; for 35 cycles. PCR products were digested with DpnI to remove template.
For myf5(80K) transformation, 1 μg of myf5(80K) DNA was used to transform competent cells, E. coli DY380. For myf5(80K) modification with GFP, putative transformants harboring myf5(80K) were isolated and grown in LB medium containing 25 μg/ml of chloramphenicol (CAM) overnight at 32°C with shaking at 150 rpm. Subsequently a 0.1% inoculum was cultured in 50 ml of LB-CAM to an OD600 of 0.8. Then, the bacteria were cultured at 42°C for 15 min, transferred to an ice slurry for 30 min, washed 5 times in ice-cold water, and electroporated immediately. Approximately 2 μg of targeting cassette DNA was mixed with the freshly prepared electrocompetent cells of DY380 carrying myf5(80K). Electroporation was performed at 1.8 kV, 200 Ohms, 25 μF for 3 × 30 s intervals (Gene Pulser Xcell, BioRad). After electroporation, cells were dispersed on the LB plates with 30 μg/ml of kanamycin and incubated at 32°C overnight. Then, recombinants were picked and checked by PCR using primers ZMFP-117F (5'-TTTGGGTGGGGATCTAGATGGTG-3') and GFP-407R (5'-GTTGCCGTCCT-CCTTGAAGT-3'), and Kan-F (5'-ATGATTGAACAAGATGGATTGC-3') and ZMF-1000R (5'-AGCGAGTTAAGTTTAAAGTCTGACCC-3') to check both integration ends.
Plasmid constructs for promoter analysis
(A) pZMYP-9977E
Following our previously described procedure [4], a 3.7-kb SacI-cut fragment from a myf5-positive recombinant bacteriophage was ligated with the SacI-digested plasmid, pZMYP-6212E, in which a -6212/-1 segment of zebrafish myf5 was fused with enhanced GFP (EGFP) cDNA. The resulting plasmid, pZMYP-9977E, contained the -9977/-1 segment of zebrafish myf5 fused with EGFP cDNA (Fig. 1B).
(B) pZMYP-9977E-ITR
A SalI/EcoRV fragment from pZMYP-6212E [4] was ligated with a Klenow-filled pGEMT-easy vector (Promega), in which the EcoRV site but not the SalI site was retained, to produce an intermediate plasmid, p(6212/1984). Then, a 3.8-kb SacI fragment from a myf5-positive phage clone [4] was ligated with SacI-treated p(6212/1984) to produce another intermediate plasmid, p(9977/1984). Finally, a SacI/EcoRV fragment cut from p(9977/1984) was ligated with SacI/EcoRV-treated pZMYP-6212E-ITR (Fig. 1B). The resulting plasmid, pZMYP-9977E-ITR, contained the -9977/-1 segment of myf5 and was flanked with inverted terminal repeats of adeno-associated virus (AAV-ITR) (Fig. 1B).
(C) pZMYP-6212E-ITR and pZMYP-2937E-ITR
A SalI/AgeI fragment from either pZMYP-6212E or -2937E [4] was ligated with a 4.2-kb SalI/AgeI fragment obtained from pCMV-EGFP-ITR [41]. The resulting plasmids, pZMYP-6212E-ITR and pZMYP-2937E-ITR, contained the -6212/-1 and -2937/-1 segments of zebrafish myf5, respectively. Each plasmid was also flanked with AAV-ITR at both ends (Fig. 1B).
(D) pZMYP-290E-ITR
Plasmid pZMYP-290E [14] was cut with HindIII, blunted and then cut with AgeI. The resulting 0.3-kb fragment was ligated with a 4.2-kb fragment produced by cutting pCMV-EGFP-ITR [41] with SalI, blunting and then cutting with AgeI. The resulting plasmid, pZMYP-290E-ITR, contained a zebrafish myf5 -290/-1 segment and was flanked with AAV-ITR at both ends (Fig. 1B).
(E) pEGFPm(2937/2457) and pEGFPm(2457/2937)
For functional identification of the notochord-specific cis-element, we used a forward primer (5'-TCTAGAACAGATTCTCATCCAA-3') and a reverse primer (5'-AACTGCACACTGGAGATTCATAAG-3') to generate module -2937/-2457. This module was ligated with pGEM T-Easy vector and then treated with EcoRI to produce the insert. EcoRI-cut pEGFPmTATA, which contained a minimal TATA-box of cytomegalovirus (CMV) promoter for the EGFP gene [14], was ligated with one copy of the EcoRI-cut module -2937/-2457 to generate pEGFPm(2937/2457). Plasmid pEGFPm(2457/2937), containing one copy of module -2457/-2937, was also constructed.
DNA preparation for microinjection and transient GFP expression
The procedures of microinjection and transient GFP detection were described by Chen et al. [4], except that we observed the GFP expression of transgenic embryos hourly, especially from 6 to 36 h.
Identification of germ-line transmission
All GFP-positive embryos at 24 h were raised to adulthood. Transgenic founders (F0) were mated with wild-types individually to confirm that they could transmit the BAC through the germ line. At least 200 embryos from each pair were examined for GFP fluorescence. After screening, GFP-positive F1 embryos were raised to adulthood and crossed with wild-type adults to generate the heterozygous F2 generation. GFP-positive F2 individuals were then crossed to each other to generate homozygous F3 fish that produced 100% GFP-positive F4 offspring.
Antibody labeling
Antibody labeling was performed as previously described, with minor modifications [42]. Embryos were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS, pH 7.0) for 4 h at room temperature, or overnight at 4°C. Then, embryos were washed in 0.1 M PBS twice for 15 min each, soaked in 100% acetone at -20°C for at least 10 min, and rehydrated with 0.1% (v/v) Tween 20 in PBS 3 times for 15 min each. After rehydration, the embryos were treated with PBS containing 5% goat serum albumin and subjected to immunofluorescence labeling. To detect zebrafish slow muscle fibers, the F59 monoclonal antibody (1:10; Hybridoma Bank) was used with Alexa Fluor 568 rabbit anti-mouse IgG (1:200; Molecular Probes) as the secondary antibody.
Whole-mount in situhybridization, cryosection and fluorescence observation
The procedures of cryosectioning and whole-mount in situ hybridization were described by Chen and Tsai [43], except that embryos from 7.5 to 60 h were used. Transgenic embryos were observed hourly, especially from 1 to 14 hpf, under a stereo fluorescence dissecting microscope (MZ12, Leica) equipped with GFP and DsRed filter cubes (Kramer Scientific). Photographs were taken with an S2 Pro digital camera (Fuji).
References
Pownall ME, Gustafsson MK, Emerson CP: Myogenic regulatory factors and the specification of muscle progenitors in verterbrate embryos. Annu Rev Cell Dev Biol. 2002, 18: 747-783. 10.1146/annurev.cellbio.18.012502.105758.
Conway K, Pin C, Kiernan JA, Merrifield P: The E protein HEB is preferentially expressed in developing muscle. Differentiation. 2004, 72: 327-340. 10.1111/j.1432-0436.2004.07207004.x.
Berkes CA, Tapscott SJ: MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005, 16: 585-595. 10.1016/j.semcdb.2005.07.006.
Chen YH, Lee WC, Liu CF, Tsai HJ: Molecular structure, dynamic expression and promoter analysis of zebrafish (Danio rerio) myf-5 gene. Genesis. 2001, 29: 22-35. 10.1002/1526-968X(200101)29:1<22::AID-GENE1002>3.0.CO;2-Z.
Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Gruwald DJ, Riggleman B: Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996, 122: 271-280.
Hadchouel J, Carvajal JJ, Daubas P, Bajard L, Chang T, Rocancourt D, Cox D, Summerbell D, Tajbakhsh S, Ribgy PWJ, Buckingham M: Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development. 2003, 130: 3415-3426. 10.1242/dev.00552.
Carvajal JJ, Cox D, Summerbell D, Rigby PWJ: A BAC transgenic analysis of the Mrf4/Myf5 locus reveals integrated elements that control activation and maintenance of gene expression during muscle development. Development. 2001, 128: 1857-1868.
Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson CP: Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev. 2002, 16: 114-126. 10.1101/gad.940702.
Buchberger A, Nomokonova N, Arnold HH: Myf5 expression in somites and limb buds of mouse embryos is controlled by two distinct distal enhancer activities. Development. 2003, 130: 3297-3307. 10.1242/dev.00557.
Mei W, Yang J, Tao Q, Geng X, Rupp RAW, Ding X: An interferon regulatory factor-like binding element restricts Xmyf 5 expression in the posterior somites during Xenopus myogenesis. FEBS Lett. 2001, 505: 47-52. 10.1016/S0014-5793(01)02688-6.
Yang J, Mei W, Otto A, Xiao L, Tao Q, Geng X, Pupp RAW, Ding X: Repression through a distal TCF-3 binding site restricts Xenopus myf5 expression in gastrula mesoderm. Mech Dev. 2002, 115: 79-89. 10.1016/S0925-4773(02)00121-1.
Lin GF, Geng X, Chen Y, Qu B, Wang F, Hu R, Ding X: T-box binding site mediates the dorsal activation of myf5 in Xenopus gastrula embryos. Dev Dyn. 2003, 226: 51-58. 10.1002/dvdy.10215.
Wang TM, Chen YH, Liu CF, Tsai HJ: Functional analysis of the proximal promoter regions of fish rhodopsin and myf5 gene using transgenesis. Mar Biotechnol. 2002, 4: 247-255. 10.1007/s10126-002-0016-y.
Chen YH, Lee HC, Liu CF, Lin CY, Tsai HJ: Novel regulatory sequence -82/-62 functions as a key element to drive the somite-specificity of zebrafish myf5. Dev Dyn. 2003, 228: 41-50. 10.1002/dvdy.10357.
Lee HC, Huang HY, Lin CY, Chen YH, Tsai HJ: Foxd3 mediates zebrafish myf5 expression during early somitogenesis. Dev Biol. 2006, 290: 359-372. 10.1016/j.ydbio.2005.11.032.
Yu D, Blis HM, Lee EC, Jenkins NA, Copeland NG, Court DL: An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000, 97: 5978-5983. 10.1073/pnas.100127597.
Lee EC, Yu D, de Velasco JM, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland G: A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting abd subcloning of BAC DNA. Genomic. 2001, 73: 56-65. 10.1006/geno.2000.6451.
Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M: Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992, 89: 8794-8797. 10.1073/pnas.89.18.8794.
Jessen RJ, Meng A, McFarlane RJ, Paw BH, Zon LI, Smith GR, Lin S: Modification of bacterial artificial chromosomes through Chi-stimulated homologous recombination and its application in zebrafish transgenesis. Proc Natl Acad Sci USA. 1998, 95: 5121-5126. 10.1073/pnas.95.9.5121.
Teboul L, Hadchouel J, Daubas P, Summerbell D, Buckingham M, Rigby PWJ: The early epaxial enhancer is essential for the initial expression of the skeletal muscle determination gene Myf5 but not for subsequent, multiple phases of somatic myogenesis. Development. 2002, 129: 4571-4580.
Teboul L, Summerbell D, Rigby PWJ: The initial somatic phase of Myf5 expression requires neither Shh signaling nor Gli regulation. Gene Dev. 2003, 17: 2870-2874. 10.1101/gad.1117603.
Lin CY, Chen YH, Lee HC, Tsai HJ: A novel cis-element in intron 1 represses somite expression of zebrafish myf5. Gene. 2004, 334: 63-72. 10.1016/j.gene.2004.03.016.
Li X, Blagden CS, Bildsoe H, Bonnin MA, Duprez D, Hughes SM: Hedgehog can drive terminal differentiation of amniote slow skeletal muscle. BMC Dev Biol. 2004, 4: 9-10.1186/1471-213X-4-9.
Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE: A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006, 10: 797-807. 10.1016/j.devcel.2006.04.009.
Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L: The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006, 20: 675-688. 10.1101/gad.1383706.
Hadchouel J, Tajbakhsh S, Primig M, Chang THT, Daubas P, Rocancourt D, Buckingham M: Modular long-range regulation of Myf5 reveals unexpected heterogeneity between skeletal muscles in the mouse embryo. Development. 2000, 127: 4455-4467.
Summerbell D, Ashby PR, Coutelle O, Cox D, Yee SP, Rigby PWJ: The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development. 2000, 127: 3745-3757.
Grove JA, Hammond CL, Hughes SM: Fgf8 drives myogenic progression of a novel lateral fast muscle fiber population in zebrafish. Development. 2005, 132: 4211-4222. 10.1242/dev.01958.
Li HY, Bourdelas A, Carron C, Gomez C, Boucaut JC, Shi DL: FGF8, Wnt8 and Myf5 are target genes of Tbx6 during anteroposterior specification in Xenopus embryo. Dev Biol. 2006, 290: 470-481. 10.1016/j.ydbio.2005.11.020.
Cossu G, Borello U: Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999, 18: 6867-72. 10.1093/emboj/18.24.6867.
Chen AE, Ginty DD, Fan CM: Protein kinase A signaling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005, 433: 208-209. 10.1038/433208a.
Shi DL, Bourdelas A, Umbhauer M, Boucaut JC: Zygotic Wnt/β-catenin signaling preferentially regulates the expression of Myf5 gene in the mesoderm of Xenopus. Dev Biol. 2002, 245: 124-135. 10.1006/dbio.2002.0633.
Coutelle O, Blagden CS, Hampson R, Halai C, Rigby PWJ, Hughes SM: Hedgehog signaling is required for maintenance of myf 5 and myoD expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev Biol. 2001, 236: 136-150. 10.1006/dbio.2001.0193.
Westerfield M: The zebrafish book. 1995, University of Oregon Press, Third
Kimmel C, Ballard WW, Kimmel SR, Ullmann B, Schilling TF: Stages of embryonic development in the zebrafish. Dev Dyn. 1995, 203: 253-310.
The Zebrafish Information Network. [http://zfin.org/zf_info/nomen.html]
RZPD German Resource Center for Genome Research. [http://www.rzpd.de]
Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu SH, Hu X, Colbert AME, Rosenberg C, Reeve-Daly MP, Rozen S, Hui L, Wu X, Vestergaard C, Wilson KM, Bae JS, Maitra S, Ganiatsas S, Evans CA, DeAngelis MM, Ingalls KA, Nahf RW, Horton LT, Anderson MO, Collymore AJ, Ye W, Kouyoumjian V, Zemsteva IS, Tam J, Devine R, Courtney DF, Renaud MT, Nguyen H, O'Connor TJ, Fizames C, Faure S, Gyapay G, Dib C, Morissette J, Orlin JB, Birren BW, Goodman N, Weissenbach J, Hawkins TL, Foote S, Page DC, Lander ES: An STS-based map of the human genome. Science. 1995, 270: 1945-1954. 10.1126/science.270.5244.1945.
LN54 panel. [http://mgchd1.nichd.nih.gov:8000/zfrh/beta.cgi]
Ensembl. [http://www.ensembl.org/Danio_rerio/]
Chou CY, Horng LS, Tsai HJ: Uniform GFP-expression in transgenic medaka (Oryzias latipes) at the F0 generation. Transgenic Res. 2001, 10: 303-315. 10.1023/A:1016671513425.
Wang YH, Chen YH, Lu CH, Tsai HJ: A 23-amino-acid motif spanning the basic domain targets zebrafish myogenic regulatory factor Myf5 into nucleolus. DNA Cell Biol. 2005, 24: 651-660. 10.1089/dna.2005.24.651.
Chen YH, Tsai HJ: Treatment with myf5-morpholino results in somite patterning and brain formation defects in zebrafish. Differentiation. 2002, 70: 447-456. 10.1046/j.1432-0436.2002.700807.x.
Acknowledgements
We are grateful to Dr. Marc Ekker for providing the LN54 mapping panel in this study. We are also grateful to Dr. Copeland, National Cancer Institute, for providing the E. coli strain DY380. This project was supported by the National Science Council, Republic of China [NSC 92-2321-B-002-018 (Y.H.C.) and NSC 92-2313-B-002-056 (H.J.T.)] and National Institutes of Health, USA [NIH HD22486 and AR45575 (M.W.)].
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
Y.H.C. designed the study, carried out the experiments, analyzed the data, and drafted the manuscript. Y.H.W. cloned the myf5-containing BAC clone, generated the Tg(myf5(80K):GFP) line, and participated in the coordination of the study and in drafting the manuscript. M.Y.C. carried out the whole-mount in situ hybridization and antibody labeling experiments. C.Y.L. generated the Tg(myf5(3K):GFP) and Tg(myf5(0.3K):GFP) lines. C.W.W. identified the notochord-restricted element. M.W and H.J.T. gave discussion and helped in editing the manuscript. H.J.T. was a principal investigator of this project.
Electronic supplementary material
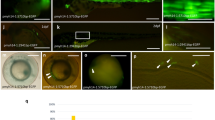
12861_2006_143_MOESM1_ESM.jpeg
Additional file 1: Cassette -2937/-2457 directs the GFP expression in notochord. Upper left: Schematic illustration of microinjected plasmids pEGFPmTATA, pEGFPm(2937/2457), and pEGFPm(2457/2937). Right: The calculation of total expression rates, notochord- and myocyte-specific expression rates, and nonspecific expression rates are described before [14]. Bottom: Embryos were photographed under fluorescent light. In pEGFPm(2937/2457)-injected zebrafish, EGFP signals appeared as bars with sharp edges (myocyte-specific) and squares (notochord-specific). Scale bar: 200 μm. (JPEG 70 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chen, YH., Wang, YH., Chang, MY. et al. Multiple upstream modules regulate zebrafish myf5expression. BMC Dev Biol 7, 1 (2007). https://doi.org/10.1186/1471-213X-7-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-213X-7-1