Abstract
Background
We developed an efficient in vitro method to differentiate mouse ES cells into the definitive endoderm (DE) and then Pdx1-expressing pancreatic lineages using mesodermal-derived supporting cells, M15. Using this method, resulting ES cell-derived DE and Pdx1-expressing cells were isolated by cell sorting, and their gene expression profiles were investigated with DNA microarray. Genes that were specifically expressed in DE and/or in Pdx1-expressing cells were extracted and their expression patterns in normal embryonic development were studied.
Results
Genes whose expression increased in DE and Pdx1 positive cells compared to the undifferentiated ES cells were chosen and in situ hybridizations were performed. Out of 54 genes examined, 27 were expressed in the DE of E8.5 mouse embryos and 15 genes were expressed in distinct domains in the pancreatic buds of E14.5 embryos. Among those genes expressed were Foxq1, CpM, Foxp4, Pcdh1, and Zmiz1, which were previously reported in other endodermal tissues. Genes, such as Parm1, Tmem184a, Hipk2 and Sox4 were reported to be expressed during early pancreatic development. Nptx2, C2cd4b, Tcf7l2 and Kiss1r were reported to be associated with beta cell or pancreatic functions in the adult. Akr1c19, Aebp2, Pbxip1 and Creb3l1, were novel and have not been described as being expressed either in DE or the pancreas.
Conclusions
We identified 27 genes, including 4 novel genes expressed in DE and pancreatic progenitor cells during normal development using an ES cell in vitro differentiation system. These results showed that DE cells and Pdx1/GFP-expressing cells obtained from our M15 based differentiation method mimic cells during the normal developmental processes. Additionally, ES cells are an excellent model for studies of early developmental processes.
Similar content being viewed by others
Background
The endoderm gives rise to the respiratory and digestive organs such as pancreas, liver, lung, stomach, and intestine. These developmental processes are of great importance in therapeutics. The multipotent endoderm has the potential to be used to repair tissues. However, in spite of the importance of the definitive endoderm (DE) derived tissues, not much is known about how they emerge from the primary gut tube. Fate mapping studies suggest that the fate of the DE begins to segregate at embryonic day 6-6.5 (E6-E6.5) and that the progenitors that are fated to become specific tissues of the gut tube appear shortly after the completion of gastrulation [1, 2]. The expression of the regional specific transcription factors has provided clues to how the endoderm is patterned into organ domains. In the chick, the progenitor for stomach, pancreas and intestine are segregated immediately after the completion of gastrulation. The progenitors receive inducing signals from the adjacent mesoderm during their migration and are specified before they reach their final destination [3]. Pancreatic and duodenal homeobox gene 1 (Pdx1) expression is by far the first sign of pancreatic differentiation detected at E8.5 in dorsal endoderm of the gut. Pdx1 is expressed before the buds become evident, and is required for the progression of pancreatic and rostral duodenal development [4]. Genetic lineage tracing studies have shown that the Pdx1-expressing cells give rise to all three cell lineages in the pancreas, the endocrine, exocrine and duct cells.
Recent advances in analysis and identification of early endodermal or pancreatic genes is remarkable [5–9]. Several reports have demonstrated the identification of novel endodermal genes using early embryos. Embryonic stem (ES) cells are also a highly useful tool in the study of endodermal development. ES cells are pluripotent cells and can be cultured indefinitely in an undifferentiated state, and stimulated to differentiate into various cell types. Progress in ES cell studies has demonstrated that ES cells provide a good system for studies of developmental biology, in addition to the use of ES cells as a surrogate cell source for regenerative medicine. Several groups have reported the differentiation of mouse or human ES cells into the DE or pancreatic lineages [10, 11]. We recently established a procedure, where ES cells were cultured on a monolayer of mesodermal-derived M15 cells, and sequentially induced into the mesendoderm, DE and regional specific DE-derived organs in vitro. This occurred in a manner that mimics early embryonic inductive events in vivo [12, 13]. With the addition of activin and bFGF, mouse ES cells differentiated into Pdx1-expressing cells efficiently, reaching 60% of the DE [12]. This M15 procedure turned out not only to be useful in directing DE lineages, but also the ectoderm and mesoderm lineages from ES cells, by altering the culture conditions [14]. Using this M15 differentiation procedure, we tried to validate the differentiated cells by analyzing the expression profiles of three germ layer cells and the pancreatic progenitor cells [14–16]. Comparison of the expression profiles between the cells of the three germ layers, which are derived from the ES cells based on this M15 procedure, revealed a clear distinct clustering of the genes specifically expressed in each germ layer. Studies of Pdx1-exprssing cells derived from ES cells led to the discovery of a novel pancreatic progenitor marker, Eppk1 [15, 17] and identification of a novel surface marker, DAF1/CD55, expressed in the DE [16]. Therefore, it is feasible to identify genes related to early DE development and pancreatic differentiation by close investigation into the ES cell-derived cells.
Here we describe an extensive gene expression profile analysis of ES cell-derived definitive endodermal cells and Pdx1-expressing cells. We chose candidate genes by the comparisons between ES cell-derived Pdx1-positive or negative DE with the undifferentiated ES cells, ES cell-derived ectoderm, mesendoderm and mesoderm cells. Then we carried out whole mount or section in situ hybridization using mouse embryos at E8.5 or pancreas at E14.5, respectively. Out of 54 candidate genes examined, 27 candidate genes are expressed in the DE at E8.5 and 15 genes are expressed in the pancreatic bud at E14.5. These results indicate that the ES cell-derived differentiated cells serve as good models for studies of candidate genes during early embryogenesis.
Results
Microarray analysis of ES cell-derived DE cell lineages
As previously described, we developed an efficient procedure using mesodermal-derived M15 cells as feeder cells with the supplementation of activin and bFGF, to sequentially differentiate ES cells into, mesendoderm (MES) at day 4 (d4), DE (E-cadherin+/CXCR4+ populations) (d5 to d7) and then Pdx1-expressing cells (d8), (Figure 1A) [12]. ES cells were also differentiated into the three germ layers under different conditions (Shiraki et al. 2009). Using these procedures, ES cell-derived differentiated cells of the ectoderm (SSEA1-/Flk1-/PGFRα-) (ECT), MES (E-cadherin+/PDGFRα+), lateral plate mesoderm (E-cadherin-/PDGFRα-/Flk1+) (LPM), paraxial mesoderm (E-cadherin-/PDGFRα+/Flk1-) (PAM) and DE at d5, d7 and d8 were prospectively isolated, by the expression of specific cell surface antigens using flow cytometry. DE cells at d8 were further subdivided into Pdx1/GFP-negative and -positive populations. The efficiencies of the induction of individual populations were similar to that previously reported (Additional file 1). The values were as follows: MES (12%; Additional file 1A), LPM (6%; Additional file 1B), PAM (42%; Additional file 1B), d5DE (45%; Additional file 1C), d5ECT (52%; Additional file 1D), d7DE (53%, Additional file 1E), d8DE (67%), and d8DE Pdx1(-) (35%x67% = 23%; Additional file 1F), or d8DE Pdx1+ (65%x67% = 42%; Additional file 1F). Since ECT was done by negative selection, we confirmed the purity of the ECT using a Sox1/GFP ES cell line [14, 18]. Ninety-four percent of the sorted ECT turned out to be Sox1/GFP-positive neuroectoderm. RNA was extracted from these cells and analyzed by Affymetrix DNA microarray. Then, gene expression profiles of undifferentiated ES cells (ES), ES cell-derived ECT, LPM, PAM, MES, d5DE, d7DE, Pdx1- d8DE and Pdx1+ d8DE, were compared. A remarkable transition of the gene expression profile was observed from d5DE to d7DE and thereafter (Figure 1B).
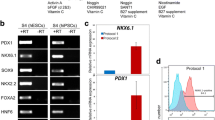
Microarray analysis of ES cell-derived cells. (A) ES cells and ES cell-derived cells were isolated by the expression of cell surface antigen as previously described [14]. Populations isolated were: ES cells (ES), ectoderm (ECT), mesendoderm (MES), lateral plate mesoderm, (LPM); Paraxial mesoderm, (PAM) and DE at day 5 (D5), day 7 (D7) and day 8 (D8, DE Pdx1-, DE Pdx1+). (B) Clustering of gene expression in ES cells, ECT, LPM, PAM, MES, d5DE, d7DE, d8DE Pdx1- and d8DE Pdx1+ cell lineages. Each line indicates an individual gene. Red lines indicate genes with high expressions and green lines indicate genes with low expressions in the DE lineages. Y axis represents normalized value of the expression level. (C) Microarray results and real time PCR analyses of six representative candidate genes, namely, AI464131, Akr1c19, Foxp4, Hipk2, Pcbd1, and Tmem184a, together with Sox17, and Pdx1, as positive controls. Right Y-axis values represent GeneChip signal intensities of candidate genes. Transcription levels of the representative candidate genes in each population are quantified by real-time PCR analysis. The transcription levels are normalized with that of b-actin. The values are further normalized with that of Pdx1-positive DE on d8 (d8DE GFP+) and thus the left Y-axis values represent relative gene expression levels when the expression level at d8 (d8DE GFP+) is defined as 100.
To validate the microarray results, we selected six representative genes, which showed increased expression levels at >7 fold changes in d8DE, compared to the median signal intensities of other populations, namely ES, ECT, LPM, PAM, MES and d5DE. We examined the actual expression patterns of these genes in ES cell-derived cells. Figure 1C shows the signal intensities of 6 genes: AI464131, Akr1c19, Foxp4, Hipk2, Pcbd1 and Tmem184a, together with those of Sox17, an endodermal gene, and Pdx1, in each indicated population of ES cell-derived cells obtained in the microarray (Figure 1C). Then, transcription levels of the above genes were quantified by real time PCR analysis (For primer sequences, see Additional file 2). The patterns of the quantitative PCR results correlated well with the signal intensities obtained in the microarray analyses (Figure 1C). The high expression level of Sox 17 in d5DE, d7DE and Pdx1 in d8DE Pdx1+ population, further confirmed the characters of the ES cell-derived cell populations.
Identification of DE specific genes in ES cell-differentiation
To identify genes involved in DE and pancreatic development, we chose genes that showed raw expression levels above 50 and increased expression in d5DE or d8DE Pdx1+ more than 5 fold change compared to the median signal intensity of ES, ECT, LPM, PAM, MES, d5DE and d8DE Pdx1+. As a result, 165 probe sets (127 genes) or 780 probe sets (594 genes) showed increased transcripts at d5DE and d8DE Pdx1+, respectively, with 115 probe sets (84 genes) overlapping between the two populations. Thus, 50 probe sets (43 genes) were specifically up regulated at d5DE, and decreased thereafter. One hundred and fifteen probe sets (84 genes) were up regulated through d5DE to d8DE Pdx1+. Six hundred and sixty-five probe sets (594 genes) were up regulated at d8DE Pdx1+.
We then examined the expression pattern of some of these DE specific genes in early stages of development. Unknown genes, or genes with specific domain structures, transcription factors or genes whose expression in the endoderm or early pancreas development have not been described, were chosen and analyzed by whole mount in situ hybridization. Figure 2 shows the summary of the numbers of genes analyzed and the genes, which were positive in the expression in the gut endoderm at embryonic day (E) E8.5 (or E9.5 for Apoe) and/or the pancreatic bud at E14.5 (Figure 2). Of the 115 probe sets (84 genes) that were up regulated at >5 fold at d5 & d8DE Pdx1+, 12 genes were picked, 8 genes were expressed in the E8.5 gut epithelium, and 4 genes were expressed in E14.5 pancreas bud (Figure 2). Of the 665 probe sets (510 genes) that were up regulated at >5 fold at d8DE, 33 genes were examined, 16 genes were expressed in the E8.5 gut epithelium, and 10 genes were expressed in the E14.5 pancreatic bud. Additionally, we randomly picked 4 genes whose expression increased at d7DE at > 5 fold compared to d5DE, and 5 genes that were expressed at d8DE in the Pdx1+ population at >2 fold compared to the Pdx1- population. Of the genes examined, 2 and 1 genes were expressed in the E8.5 gut epithelium, respectively, and 2 (1+1) genes were expressed in E14.5 pancreas bud (Figure 2).
Numbers of endoderm specific candidate genes. Summary of the numbers of genes picked up for further analyses by whole mount in situ hybridization (top). Numbers of genes that are expressed in gut endoderm at E8.5 (middle) or the pancreatic bud at E14.5 (bottom). Blue circle: genes expressed at >5 fold in d5DE; Red circle: genes expressed at >5 fold in d8DE Pdx1+; Green circle: genes expressed at >5 fold in d7DE versus d5DE; Brown circle, genes expressed at >2 fold in d8DE Pdx1+ versus d8DE Pdx1-
Genes up regulated in ES cell-derived DE cells are also expressed in the gut endoderm at E8.5
As shown in Figure 2, out of 12 genes examined, the expression of 8 genes, which increased at >5 fold at d5DE through d8DE, were expressed in E8.5 or E9.5 endoderm. The 8 positive genes were: AI464131, Akr1c19, DAF1/CD55, Foxq1, Nptx2, Pga5, Parm1 and Tmem184a. Their expression patterns at E8.5 are shown in Figure 3. AI464131 and Nptx2 were expressed in the whole gut endoderm throughout the anterior-to-posterior region (Figure 3). Akr1c19 was expressed weakly in the anterior intestinal portal (AIP, depicted by an arrow) at E8.5, and in the liver bud, pancreas and intestine epithelium at E9.5 (SH data not shown). DAF1/CD55 expression is observed in the lateral gut and AIP at E8.5, which was published earlier [16] (Table 1). Similarly, Tmem184a was expressed in the lateral gut and AIP at E8.5. Foxq1 was expressed strongly in the AIP. Parm1 was expressed in the dorsal anterior gut epithelium. Pga5 was expressed weakly in the whole lateral gut epithelium. The regions positively detected by in situ hybridization are marked by arrows (Figure 3). Cross sections were made and representative results of Parm1 and Tmem184a expressions are shown (Figure 3).
The expression pattern of 7 candidate genes, whose expression is increased at d5DE in E8.5 mouse definitive endoderm, analyzed by in situ hybridization. In situ hybridization pictures of AI464131, Akr1c19, Foxq1, Pga5, Parm1, Nptx2 and Tmem184a are shown. Arrows indicate the signals detected. Cross sections through the white line of Parm1 and Tmem184a are shown (right panels). A: anterior; P: posterior. Scale bars: 100 μm
The expression patterns by in situ hybridization in E8.5 (or E9.5 with Apoe gene) of 16 positive genes, whose expression increased first at d8DE are shown in Figure 4A. Apoe was expressed in the lateral endoderm and visceral endoderm at E8.5 (SH unpublished) and strongly in the liver bud at E9.5 (Figure 4A, Apoe, arrow). Most genes, including Chi3l1, CpM, C2cd4b, Fam184b, Fhl2, Foxp4, Hipk2, Lbh, Pcbd1, Pbxip1, Sox4, Tcf7l2 and Zmiz1, were expressed in the anterior intestinal portal (AIP) in E8.5 embryos (Figure 4A, arrows). In addition to the AIP, C2cd4b was also expressed in the hindgut; CpM, Foxp4 and Zmiz were also expressed in the lateral gut epithelium. Irf6 was expressed in the anterior endoderm and hindgut epithelium. The lateral gut epithelium expression of Zmiz1 was shown by sectioning the E8.5 embryo after in situ hybridization (Figure 4A, arrows). An Ig domain gene, Palld1, was expressed in the dorsal gut epithelium, also confirmed by sectioning the E8.5 embryo after in situ hybridization (Figure 4A, arrow).
The expression pattern of 18 candidate genes, whose expression is increased, at d8DE, in E8.5 mouse definitive endoderm, analyzed by in situ hybridization. (A) The expression of 16 candidate genes, whose expression is increased >5 fold in d8DE Pdx1/GFP+ population, (B) >5 fold in d7DE versus d5DE, or (C) >2 fold in d8DE Pdx1+ versus Pdx1- cells, are shown. Gene names are indicated at top of the pictures. Arrows indicate the sites detected. Cross sections through the white line of Palld1 or Zmiz1 are shown (right panels). A: anterior; P: posterior. Scale bars: 100 μm
The followings are the expression patterns of the genes that were up regulated at d7DE or d8DE, at >5 fold compared to d5DE (Figure 4B) or at a >2 fold increase in Pdx1+ versus Pdx1- population (Figure 4C): Among the 9 genes examined (Figure 2B, Table 1), Aebp2, Barhl and Kiss1r were expressed in E8.5 embryos. Aebp2 was strongly expressed in the whole gut tube, and Barhl2, Kiss1r in the lateral gut epithelium (Figure 4B,C, arrows).
All genes described in Figure 3 and 4 are summarized in Table 1, with their gene descriptions, Genbank number, expression patterns in E8.5 endoderm, and previous publications on expression or function in the endoderm or pancreas. Although they have been implicated in the function of pancreas or other endodermal derived organs, this study showed, for the first time, the expression of many genes in the early stage of development.
Genes expressed in distinct pancreas domains at E14.5
As described above, of the 54 genes analyzed, 27 genes were expressed in the E8.5 gut epithelium (Figure 3, 4). It was reported that transcripts of genes could be categorized based on their expression patterns into one of five expression domain in the pancreas at E14.5 [5]. We then examined all 54 genes for their expression in E14.5 pancreatic buds. Among the 54 genes, 15 genes are expressed in E14.5 pancreatic buds. The expression patterns were categorized (Figure 5, 6, Table 1). Aebp2, AI464131, Akr1c19, Creb3l1, Foxp4, Hipk2, Pcbd1, Sox4, Tcf7l2 and Tmem184a were expressed in the pancreas epithelium (Figure 5). Pbxip1 and C2cd4b were expressed in the trunk region (Figure 6A). Parm1 and Pcdh1 were expressed in the tip region (Figure 6B). AI464131, Creb3l1 and Tcf7l2 were also expressed in the mesenchyme, in addition to their epithelial expressions (Figure 6C). Pcdh1 was expressed only in the mesenchyme (Figure 6C). Apoe was expressed in the vascular cells (Figure 6D). The above gene expression domains in E14.5 pancreatic buds are summarized in Table 1.
The expression of 10 candidate genes in E14.5 pancreas epithelium. Aebp2, AI464131, Akr1c19, Creb3l1, Foxp4, Hipk2, Pcbd1, Sox4, Tcf7l2 and Tmem184a are expressed in the pancreas epithelium, detected by in situ hybridization. Parts of the pancreas epithelia are out lined with dotted lines. Scale bar: 100 μm, applied to all pictures.
The expression of candidate genes in E14.5 pancreas distinct regions. (A) Pbxip1 and C2cd4b are expressed in the trunk region. (B) Parm1 and Pcdh1 are in the tip region. (C) AI464131, Creb3l1, Pcdh1 and Tcf7l2 are expressed in the mesenchyme. Pcdh1 is expressed exclusively in the mesenchyme, whereas the others are also expressed in the epithelium. (D) Apoe is expressed in the vascular cells. Parts of the pancreas epithelia are out lined with dotted line in A-C. Scale bars: 100 μm
Coexpression of the genes with endocrine and exocrine markers in the pancreatic bud
To investigate further into the function of the genes, their co-expression with the endocrine markers, namely insulin and glucagon, or an exocrine marker, amylase, and Pdx1 were examined.
C2cd4b, a gene expressed in the trunk (Figure 7A), was co-expressed with insulin, but not glucagon, implicating its function in endocrine β cell differentiation (Figure 7A). Hipk2 was co-expressed with glucagon, but not insulin, implicating that it might associate with α cell differentiation (Figure 7A). Akr1c19 was co-expressed with Pdx1 or insulin, thus suggesting its possible function in β cell differentiation (Figure 7A). Foxp4, Pcbd1 and Aebp2, which are expressed in the epithelium, were co-localized in part with amylase staining (Figure 7B).
The co-expression of candidate genes with insulin, glucagon, Pdx1 or amylase in E14.5 pancreatic bud. (A) C2cd4b co-expressed with insulin but not glucagon in the trunk (Upper panels). Hipk2 co-expressed with glucagon but not insulin in the epithelium. Akr1c19 co-expressed with Pdx1 or insulin in the epithelium. (B) Foxp4, Pcbd1 and Aebp2 partly co-expressed with amylase. Scale bar: 100 μm, applied to A-B.
Discussion
We previously reported the prospective isolation and global expression profiles of the ES cells derived three germ layer cells, such as the mesendoderm, ectoderm, mesoderm and DE, which were obtained by culturing ES cells on M15 [14]. Therefore, M15 provides a platform to compare expression profiles of different lineages derived from ES cells.
In our previous analyses, without the addition of activin and bFGF, but with M15 feeder cells, ES cells derived into immature DE cells, and there were few genes that were expressed in E14.5 pancreatic buds [15]. Here, we used a differentiation protocol with the supplementation of activin and bFGF, a procedure that allows for approximately 50-60% of the differentiated ES cells to give rise to the DE, and among which 60% cells give rise to Pdx1-expressing cells [12, 14]. Under the present protocol, ES cells differentiated into a pancreatic developmental pathway. Many genes expressed specifically in the pancreas are identified by comparing d7DE or d8DE with other populations. This may explain the reason for the small differences between GFP+ and GFP- population, since GFP- population are committed to a similar developmental pathway. Using this protocol, we efficiently identified genes specifically expressed in the DE and the pancreatic lineage.
We also examined in our ES cell-derived cells, the expression levels of genes enriched the E7.5 embryo endoderm (Additional file 3) [19], E8.25 endoderm (Additional file 4) [7], and E10.5 Pdx1+ cells (Additional file 5) [19]. Many of the enriched genes in DE of E7.5 embryos were also found to be up-regulated in our ES cell-derived d5DE, d7DE and d8DE GFP+ cells (Additional file 3). Of the endoderm enriched genes in E8.25 embryos, genes such as Spink3, Clic6, FoxA1, Krt7, Crb3, Ell3, Rab15 Rbm35a, St14 and Tmprss2 are expressed at a higher level in d7DE or d8DE than d5DE. A limited number of E10.5 Pdx1+ cells enriched genes, such as Mest, Peg3, Col1a2, Tnc, Capn, Gap43 and Meis1, were up regulated in the d8DE GFP+ cells. These results therefore suggest that gene expression profiles in d5, d7 and d8DE were similar with that of E7.5 embryonic DE. However, d7 and d8DE resembled E8.25 embryonic DE compared to d5DE, whereas d8DE resembled E10.5 embryonic Pdx1+ cells compared to the other populations.
Genes expressed in endodermal tissues other than the pancreas
Among the genes whose expression is increased in the ES cell-derived DE population, several genes are found to be expressed in the foregut, hindgut or whole gut. There are genes previously reported to be expressed in the gut endoderm, or genes with some aspects related to pancreas endocrine or exocrine functions.
Foxq1, a fork-head transcription factor, was previously described to be specifically expressed in the stomach in developing and adult gastrointestinal tract. Mice carrying Foxq1 mutations show absence of mRNA and protein of the major stomach mucin MUC5AC, and the mutant mice show a paucity of foveolar cell secretory vesicles and a notable loss of stomach mucus [20].
CpM, Carboxypeptidase M, is a glycosylphosphatidylinositol (GPI)-anchored membrane protein with B-type carboxypeptidase activity and is a member of the 'regulatory' or carboxypeptidase N/E subfamily of metallocarboxypeptidases. CpM expression has been described in the developing and adult lung in Alveolar type I cells [21, 22]. Recently, the expression of CpM was described to be one of the genes expressed in the Foxa2-expressing endoderm cells [23].
Foxp4, a member of the forkhead box family of winged-helix genes, is described to be expressed in the pulmonary, neural and gut tissues during embryonic development [24]. It was recently reported to mediate transcriptional repression, by interaction with a component of NuRD, and regulate gene expression and epithelial injury response in the lung via regulation of interleukin-6 [25].
Pcdh1, Protocadherin-1, is a member of the δ-protocadherin subgroup of non-clustered protocadherins. The expression is described to be especially prominent in blood vessels in organs derived from the embryonic gut, such as the esophagus, intestines, liver, lung, and submandibular glands [26].
Zmiz1, also known as Zmip10, is a member of PIAS (protein inhibitor of activated STAT) family of genes. Like the other members of this family, Zmiz1 contains the zinc-binding SP-RING Zn-finger domain, which confers SUMO-conjugating activity. At E7.5 Zmiz1 transcripts were generally observed in the epiblast, and expression was detected in the neuroectoderm, dorsal aorta and foregut at E8.5 [27]. Taken together, these results suggest that genes up regulated in the ES cell-derived DE at d5 and later, or genes up regulated at d8, are good candidates for being expressed in the gut endoderm at E8.5 during early embryogenesis. Therefore, ES cell-derived DE resembles normal early embryos, and serves as a good source for providing cells for studies on development.
Genes expressed in the pancreas
Since the differentiation procedure for DE at d7 or d8 is in favor of pancreatic differentiation, there turned out to be many genes expressed in the pancreatic bud. Parm1, Riken cDNA 9130213B05, is one of the top ranked 73 genes whose expression is decreased in Pdx1 mutant mice in the E10.5 dorsal pancreatic bud [6]. Tmem184, also described as Sdmg1, is a component of secretory granules in mouse secretory exocrine tissues, such as pancreas, salivary gland, and mammary gland. Its expression in the developing pancreas was recently reported [28]. homeodomain-interacting protein kinas (HIPK)-2, is expressed in the embryonic and adult pancreas and positively regulates Pdx1 transcriptional activity by directly phosphorylating the C-terminal portion of Pdx1 [29]. Nptx2 is one of genes reported with aberrant DNA methylation in familial pancreatic cancers [30].
Tcf7l2, also known as TCF-4, is a component of the canonical Wnt signaling pathway. A strong genetic association relationship between Tcf7l2 polymorphisms and type 2 diabetes mellitus was identified [31]. Tcf7l2 was shown to be involved in stimulating the proliferation of pancreatic β-cells and the production of the incretin hormone glucagon-like peptide-1 in intestinal endocrine L cells [32]. Silencing of TCF7L2 in human and mouse islets results in a decrease in glucose-stimulated insulin secretion and increased beta cell apoptosis [33, 34].
Similarly, variants at C2cd4b loci are associated with reduced glucose-stimulated β cell function [35]. Sox4, a gene of a member of the homeodomain and basic helix-loop-helix families of transcription factors, is expressed in developing mouse pancreas [36]. Sox4 reportedly has a role in insulin secretion in the adult β-cell downstream of the KATP channel, and is identified as a risk factor for diabetes and obesity [37, 38]. Kiss1r, also known as GPR54, is expressed in the islets and in β (MIN5) and α (alphaTC1) cell lines [39]. Kisspeptin, a novel peptide as endogenous ligand of Kiss1r, plays a crucial role in puberty and reproductive function [40], and potentiates glucose-induced insulin secretion from isolated islets [41].
Most genes listed here are revealed for the first time to be expressed at this early stage of E8.5 and E14.5. It is of interest that genes responsible for β-cell maturation are expressed at early stages of development. Future works examining their function would reveal their role in replication or differentiation of the pancreas. These results again highlight the usefulness of the ES cell-derived cells for the discovering of genes that have a role in the development and function of the pancreas.
Genes found for the first time to be expressed in the definitive endoderm or pancreas
There are genes that have never reported to be expressed in the DE or the pancreas. Akr1c19 is an aldo-keto reductase family 1 member C19 [42]. Aebp2 encodes a zinc finger protein that has been shown to interact with the mammalian Polycomb Repression Complex2 (PRC2) [43]. Its Drosophila homolog, jing, is a zinc-finger transcription factor that interacts with the fly Polycomb Group (PcG) protein complexes, and plays an essential role in controlling CNS midline and tracheal cell differentiation [44]. Pbxip1 is a PBX interacting protein, also known as HPIP, inhibits the binding of Pbx1-Hox complexes to DNA [45]. Creb3l1, also known as OASIS, is a ZIP (basic leucine zipper) transcription factor, which is a member of the CREB/ATF family, has a transmembrane domain, and has been identified as an ER stress transducer [46]. The physiological functions of these genes in normal pancreatic development remain to be explored.
Finally, there are genes whose expression we could not detect in the Pdx1+ cells during normal pancreatic development. This might be due to their low expression levels and/or technical limitation. In addition, some of the genes show expression patterns that are difficult to be catalogued at E14.5, since the pancreatic differentiation undergoes a secondary transition at this stage and many genes show a dramatic change in their expression patterns after the secondary transition.
Conclusions
Taken together, our data show that ES cell in vitro differentiation is an excellent model system for studies of early developmental processes. ES cell-derived DE cells or Pdx1/GFP-expressing cells obtained from M15 based differentiation procedure mimic cells of the normal developmental processes. Markers identified will be useful for the elucidation of the pancreatic development. This data will also help to derive cells from ES cells that are useful for therapeutics.
Methods
Cell lines
The mouse ES cell line, SK7, containing a Pdx1 promoter-driven GFP reporter transgene, and the mesonephric M15 cells were cultured and differentiated as previously described [12–14, 47].
Differentiation of ES cells
For mesendoderm (MES), definitive endoderm (DE) differentiation and pancreatic differentiation, ES cells were cultured for 4, 5 or 8 days on M15 added with 20 ng/ml recombinant human activin-A and 50 ng/ml human bFGF, respectively. For neuroectoderm (ECT) differentiation, ES cell culture on M15 was supplemented with 10 μM SB203580 (a MAPK inhibitor). For lateral plate mesoderm (LPM) or paraxial mesoderm (PAM) differentiation, ES cell culture on M15 was supplemented with BMP7 at 25 ng/ml. Isolation of ES cell-derived ECT, MES, and mesoderm (PAM+LPM) cells was performed as previously described [14]. Briefly, MES, DE and mesoderm were isolated as E-cadherin+/PDGFRα+, E-cadherin+/Cxcr4+, and E-cadherin- and Flk1+ or PDGFRα+ fraction, respectively, by cell sorting. Ectoderm cells were isolated as PDGFRα-/Flk1-/SSEA1-. The result of ectoderm differentiation using Sox1/GFP ES cells indicated that over 90% of sorted PDGFRα-/Flk1-/SSEA1- cells grown on M15 cell with SB203580 are Sox1-positive neuroectoderm cells (Additional file 1).
Flow cytometry analysis
Cells were dissociated with Cell Dissociation Buffer (Invitrogen, Carlsbad, CA) and adjusted to 1 × 106 cells/50 μl and stained with appropriate antibodies. The following antibodies were used: biotin- or Alexa 488-conjugated anti-E-cadherin monoclonal antibody (mAb) ECCD-2, biotin- or phycoerythrin (PE)-conjugated anti-CXCR4 mAb 2B11 (BD Pharmingen, San Diego, CA), PE-conjugated anti-SSEA1 mAb (R&D Systems), PE-conjugated anti-FLK1 mAb AVAS12 (BD Biosciences Pharmingen, San Diego, CA), biotin-conjugated anti-PDGFRα mAb APA5 (BD Pharmingen) and streptavidin-allophycocyanin (BD Pharmingen). The stained cells were analyzed using a FACS Canto (BD Pharmingen) or purified with FACS Aria (BD Pharmingen). Data were recorded using the BD FACSDiva Software program (BD Pharmingen) and analyzed using the Flowjo program (Tree Star, Ashland, OR).
Growth factors and inhibitors
The following concentrations were used: recombinant human activin-A (R&D Systems), 20 ng/ml. Human bFGF (Peprotech), 50 ng/ml; recombinant human BMP7 (R&D Systems), 25 ng/ml; SB203580 (Calbiochem), 10 μM.
Microarray analysis
The biotinylated cRNAs from differentiated ES cells were hybridized with a MOE430 2.0 series probe array (GeneChip, Affymetrix). The fluorescence intensity of each probe was quantified using the GeneChip Analysis Suite 5.0 computer program (Affymetrix). Data from individual arrays were normalized and expression analysis was performed using the GeneSpring GX software program version 7.3 (Agilent).
Quantitative RT (Reverse transcription) - real time PCR(polymerase chain reaction)
RNA was extracted from ES cells using TRI Reagent (Sigma) or RNeasy micro-kit (Qiagen) and then treated with DNase (Sigma). Three micrograms of RNA was reverse-transcribed using MMLV reverse transcriptase (Toyobo) and oligo dT primers (Toyobo). The primer sequences are shown in Additional file 2. The transcript was quantified with SyberGreen on an ABI 7500 thermal cycler (Applied Biosystems). The PCR conditions were as follows: denaturation at 95°C for 15 sec, annealing and extention at 60°C for 60 sec, for up to 40 cycles. Transcripts were normalized to ß-actin by subtracting the average ß-actin Ct values (Threshold Cycle) from the average Ct of transcripts, resulting in Ct. Target mRNA levels were determined by standard curve method and expressed as arbitrary units.
In situ hybridization
For whole mount in situ hybridization, embryos were fixed with 4% paraformaldehyde overnight, dehydrated in methanol, treated with proteinase K at 10 μg/ml for 10 min, and then refixed in 0.2% glutaraldehyde/4% paraformaldehyde. Hybridization were performed with Digoxigenin (DIG) (Roche) labeled antisense RNA probes at 1 μg/ml. Plasmids for RNA probes were purchased from Open Biosystems or RIKEN BioResource Center (Tsukuba).
For in situ hybridization on sections, embryos were fixed with 4% paraformaldehyde overnight, replaced with 30% sucrose in PBS 4˚C overnight and embedded in OCT compound (Sakura Fine technical Co.). In situ hybridization with digoxigenin-labeled probes was performed on 10 μm frozen section as described [48, 49].
Immunocytochemistry
The following antibodies were used: goat anti-Amylase (Santa Cruz Biotechnology Inc., Santa cruz, CA), rabbit anti-GFP (MBL International Corp., Woburn, MA), mouse anti-insulin (Sigma-Aldrich), guinea pig anti-glucagon (Linco Research., St.Charles, MO).
References
Tremblay KD, Zaret KS: Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol. 2005, 280: 87-99. 10.1016/j.ydbio.2005.01.003.
Lawson KA, Meneses JJ, Pedersen RA: Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991, 113: 891-911.
Katsumoto K, Shiraki N, Miki R, Kume S: Embryonic and adult stem cell systems in mammals: ontology and regulation. Dev Growth Differ. 2010, 52: 115-29. 10.1111/j.1440-169X.2009.01160.x.
Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV: PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996, 122: 983-95.
Zhou Q, AC Law, Rajagopal J, Anderson WJ, Gray PA, Melton DA: A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007, 13: 103-14. 10.1016/j.devcel.2007.06.001.
Svensson P, Williams C, Lundeberg J, Ryden P, Bergqvist I, Edlund H: Gene array identification of Ipf1/Pdx1-/- regulated genes in pancreatic progenitor cells. BMC Dev Biol. 2007, 7: 129-10.1186/1471-213X-7-129.
Sherwood RI, Jitianu C, Cleaver O, Shaywitz DA, Lamenzo JO, Chen AE, Golub TR, Melton DA: Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev Biol. 2007, 304: 541-55. 10.1016/j.ydbio.2007.01.011.
Sherwood RI, Chen TY, Melton DA: Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009, 238: 29-42. 10.1002/dvdy.21810.
Hoffman BG, Zavaglia B, Witzsche J, Ruiz de Algara T, Beach M, Hoodless PA, Jones SJ, Marra MA, Helgason CD: Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol. 2008, 9: R99-10.1186/gb-2008-9-6-r99.
D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE: Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006, 24: 1392-401.
KA D'Amour, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE: Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005, 23: 1534-41. 10.1038/nbt1163.
Shiraki N, Yoshida T, Araki K, Umezawa A, Higuchi Y, Goto H, Kume K, Kume S: Guided differentiation of embryonic stem cells into Pdx1-expressing regional-specific definitive endoderm. Stem Cells. 2008, 26: 874-85. 10.1634/stemcells.2007-0608.
Shiraki N, Umeda K, Sakashita N, Takeya M, Kume K, Kume S: Differentiation of mouse and human embryonic stem cells into hepatic lineages. Genes Cells. 2008, 13: 731-46. 10.1111/j.1365-2443.2008.01201.x.
Shiraki N, Higuchi Y, Harada S, Umeda K, Isagawa T, Aburatani H, Kume K, Kume S: Differentiation and characterization of embryonic stem cells into three germ layers. Biochem Biophys Res Commun. 2009, 381: 694-9. 10.1016/j.bbrc.2009.02.120.
Yoshida T, Murata K, Shiraki N, Kume K, Kume S: Analysis of gene expressions of embryonic stem-derived Pdx1-expressing cells: implications of genes involved in pancreas differentiation. Dev Growth Differ. 2009, 51: 463-72. 10.1111/j.1440-169X.2009.01109.x.
Shiraki N, Harada S, Ogaki S, Kume K, Kume S: Identification of DAF1/CD55, a novel definitive endoderm marker. Cell Struct Funct. 2010, 35: 73-80. 10.1247/csf.10004.
Yoshida T, Shiraki N, Baba H, Goto M, Fujiwara S, Kume K, Kume S: Expression patterns of epiplakin1 in pancreas, pancreatic cancer and regenerating pancreas. Genes Cells. 2008, 13: 667-78. 10.1111/j.1365-2443.2008.01196.x.
Ying QL, Nichols J, Chambers I, Smith A: BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003, 115: 281-92. 10.1016/S0092-8674(03)00847-X.
Gu G, Wells JM, Dombkowski D, Preffer F, Aronow B, Melton DA: Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development. 2004, 131: 165-79. 10.1242/dev.00921.
Verzi MP, Khan AH, Ito S, Shivdasani RA: Transcription factor foxq1 controls mucin gene expression and granule content in mouse stomach surface mucous cells. Gastroenterology. 2008, 135: 591-600. 10.1053/j.gastro.2008.04.019.
Nagae A, Abe M, Becker RP, Deddish PA, Skidgel RA, Erdos EG: High concentration of carboxypeptidase M in lungs: presence of the enzyme in alveolar type I cells. Am J Respir Cell Mol Biol. 1993, 9: 221-9.
Fujiwara N, Ikeda M, Hirabayashi S, Mori H, Shirasawa M, Kansaku A, Sunamori M, Hata Y: Monoclonal antibody 7F9 recognizes rat protein homologous to human carboxypeptidase-M in developing and adult rat lung. Respirology. 2007, 12: 54-62. 10.1111/j.1440-1843.2006.00961.x.
Tamplin OJ, Kinzel D, Cox BJ, Bell CE, Rossant J, Lickert H: Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics. 2008, 9: 511-10.1186/1471-2164-9-511.
Lu MM, Li S, Yang H, Morrisey EE: Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech Dev. 2002, 119 (Suppl 1): S197-202. 10.1016/S0925-4773(03)00116-3.
Chokas AL, Trivedi CM, Lu MM, Tucker PW, Li S, Epstein JA, Morrisey EE: Foxp1/2/4-NuRD interactions regulate gene expression and epithelial injury response in the lung via regulation of interleukin-6. J Biol Chem. 285: 13304-13. 10.1074/jbc.M109.088468.
Redies C, Heyder J, Kohoutek T, Staes K, Van Roy F: Expression of protocadherin-1 (Pcdh1) during mouse development. Dev Dyn. 2008, 237: 2496-505. 10.1002/dvdy.21650.
Rodriguez-Magadan H, Merino E, Schnabel D, Ramirez L, Lomeli H: Spatial and temporal expression of Zimp7 and Zimp10 PIAS-like proteins in the developing mouse embryo. Gene Expr Patterns. 2008, 8: 206-13. 10.1016/j.modgep.2007.10.005.
Best D, Adams IR: Sdmg1 is a component of secretory granules in mouse secretory exocrine tissues. Dev Dyn. 2009, 238: 223-31. 10.1002/dvdy.21827.
Boucher MJ, Simoneau M, Edlund H: The homeodomain-interacting protein kinase 2 regulates insulin promoter factor-1/pancreatic duodenal homeobox-1 transcriptional activity. Endocrinology. 2009, 150: 87-97. 10.1210/en.2007-0865.
Brune K, Hong SM, Li A, Yachida S, Abe T, Griffith M, Yang D, Omura N, Eshleman J, Canto M, et al: Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiol Biomarkers Prev. 2008, 17: 3536-42. 10.1158/1055-9965.EPI-08-0630.
Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al: Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006, 38: 320-3. 10.1038/ng1732.
Jin T, Liu L: The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol. 2008, 22: 2383-92. 10.1210/me.2008-0135.
Shu L, Sauter NS, Schulthess FT, Matveyenko AV, Oberholzer J, Maedler K: Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008, 57: 645-53. 10.2337/db07-0847.
da Silva Xavier G, Loder MK, McDonald A, Tarasov AI, Carzaniga R, Kronenberger K, Barg S, Rutter GA: TCF7L2 regulates late events in insulin secretion from pancreatic islet beta-cells. Diabetes. 2009, 58: 894-905. 10.2337/db08-1187.
Boesgaard TW, Grarup N, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O: Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia. 2010, 53: 1647-55. 10.1007/s00125-010-1753-5.
Lioubinski O, Muller M, Wegner M, Sander M: Expression of Sox transcription factors in the developing mouse pancreas. Dev Dyn. 2003, 227: 402-8. 10.1002/dvdy.10311.
Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes O, Engstrom PG, Alonso ME, de la Calle Mustienes E, Gomez Skarmeta JL, Tavares MJ, et al: Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci USA. 2010, 107: 775-80. 10.1073/pnas.0911591107.
Goldsworthy M, Hugill A, Freeman H, Horner E, Shimomura K, Bogani D, Pieles G, Mijat V, Arkell R, Bhattacharya S, et al: Role of the transcription factor sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008, 57: 2234-44. 10.2337/db07-0337.
Hauge-Evans AC, Richardson CC, Milne HM, Christie MR, Persaud SJ, Jones PM: A role for kisspeptin in islet function. Diabetologia. 2006, 49: 2131-5. 10.1007/s00125-006-0343-z.
Funes S, Hedrick JA, Vassileva G, Markowitz L, Abbondanzo S, Golovko A, Yang S, Monsma FJ, Gustafson EL: The KiSS-1 receptor GPR54 is essential for the development of the murine reproductive system. Biochem Biophys Res Commun. 2003, 312: 1357-63. 10.1016/j.bbrc.2003.11.066.
Bowe JE, King AJ, Kinsey-Jones JS, Foot VL, Li XF, O'Byrne KT, Persaud SJ, Jones PM: Kisspeptin stimulation of insulin secretion: mechanisms of action in mouse islets and rats. Diabetologia. 2009, 52: 855-62. 10.1007/s00125-009-1283-1.
Ishikura S, Horie K, Sanai M, Matsumoto K, Hara A: Enzymatic properties of a member (AKR1C19) of the aldo-keto reductase family. Biol Pharm Bull. 2005, 28: 1075-8. 10.1248/bpb.28.1075.
Kim H, Kang K, Kim J: AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009, 37: 2940-50. 10.1093/nar/gkp149.
Sedaghat Y, Miranda WF, Sonnenfeld MJ: The jing Zn-finger transcription factor is a mediator of cellular differentiation in the Drosophila CNS midline and trachea. Development. 2002, 129: 2591-606.
Abramovich C, Shen WF, Pineault N, Imren S, Montpetit B, Largman C, Humphries RK: Functional cloning and characterization of a novel nonhomeodomain protein that inhibits the binding of PBX1-HOX complexes to DNA. J Biol Chem. 2000, 275: 26172-7. 10.1074/jbc.M001323200.
Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, et al: Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009, 11: 1205-11. 10.1038/ncb1963.
Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND: Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995, 81: 391-401. 10.1016/0092-8674(95)90392-5.
Matsuura K, Katsumoto K, Fukuda K, Kume K, Kume S: Conserved origin of the ventral pancreas in chicken. Mech Dev. 2009, 126: 817-27. 10.1016/j.mod.2009.07.009.
Katsumoto K, Fukuda K, Kimura W, Shimamura K, Yasugi S, Kume S: Origin of pancreatic precursors in the chick embryo and the mechanism of endoderm regionalization. Mech Dev. 2009, 126: 539-51. 10.1016/j.mod.2009.03.006.
Acknowledgements
We thank members of the Gene Technology Center and the Center for Animal Resources and Development at Kumamoto University for their technical assistance. S.K. is a global COE member. This work was supported by a Grant-in-Aid (#21390280 to S.K. and #21790671 to N.S.), and in part by a Global COE grant (Cell Fate Regulation Research and Education Unit), a grant (to S.K.) from the Project for Realization of Regenerative Medicine from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SO and SH contributed to acquisition, analysis and interpretation of data. NS contributed to the conception and design, analysis and interpretation of data, and drafting the manuscript. KK carried out interpretation of data, and drafting the manuscript. SK contributed to the conception and design, interpretation of data, financial supports, and drafting the manuscript. All authors critically read, revised and approved the final manuscript.
Soichiro Ogaki, Seiko Harada contributed equally to this work.
Electronic supplementary material
12861_2010_612_MOESM1_ESM.JPEG
Additional file 1: Flow cytometric analyses of ES cell-derived cells of each cell population. (A) D4 MES (E-cadherin-/PDGFRα+). (B) D5 Mesoderm: PAM (E-cadherin-/PDGFRα+/Flk1-) and LPM (E-cadherin-/PDGFRα+/Flk1-). (C) D5DE (E-cadherin+/CXCR4+), (D) D5 ECT (SSEA1-/Flk1-/PGFRα-), then sorted with Sox1/GFP (X-axis), plotted against FSC (Forward scatter; Y-axis). (E) D7DE (E-cadherin+/CXCR4+), (F) D8DE (E-cadherin+/CXCR4+), then sorted with Pdx1/GFP (X-axis), plotted against E-cadherin (Y-axis). Cells collected as the indicated cell population are shown in blue squares (A, left panel in B, C-F), or in the quadrants (right panel in B). The percentages of the collected cell populations are displayed. (JPEG 374 KB)
12861_2010_612_MOESM3_ESM.PDF
Additional file 3: A comparisons with E7.5 endoderm enriched genes (versus other germ layers) (Gu et al., Development, 2004). (PDF 53 KB)
12861_2010_612_MOESM4_ESM.PDF
Additional file 4: A comparison with E8.25 endoderm enriched genes (Sherwood et al., Developmental biology, 2007). (PDF 48 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ogaki, S., Harada, S., Shiraki, N. et al. An expression profile analysis of ES cell-derived definitive endodermal cells and Pdx1-expressing cells. BMC Dev Biol 11, 13 (2011). https://doi.org/10.1186/1471-213X-11-13
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-213X-11-13