Abstract
Background
Expression of transgenes in mice requires transcriptional regulatory elements that direct expression in a chosen cell type. Unfortunately, the availability of well-characterized promoters that direct bona-fide expression of transgenes in transgenic mice is limited. Here we described a method that allows highly efficient targeting of transgenes to a preselected locus in ES cells.
Results
A pgk-LoxP-Neo cassette was introduced into a desired genomic locus by homologous recombination in ES cells. The pgk promoter was then removed from the targeted ES cells by Cre recombinase thereby restoring the ES cells' sensitivity to G418. We demonstrated that transgenes could be efficiently introduced into this genomic locus by reconstituting a functional Neo gene.
Conclusion
This approach is simple and extremely efficient in facilitating the introduction of single-copy transgenes into defined genomic loci. The availability of such an approach greatly enhances the ease of using endogenous regulatory elements to control transgene expression and, in turn, expands the repertoire of elements available for transgene expression.
Similar content being viewed by others
Introduction
The study of gene function has been greatly advanced through the use of transgenic mice. Over-expression and ectopic expression of transgenes, as well as the expression of genetic variants, has facilitated the deciphering of complex cellular processes as well as the generation of animal models of disease. More recently, transgenic mice have also been used to express Cre recombinase to mediate the conditional deletion of loxP-flanked DNA sequences in specific cell types in vivo. Traditional methods for generating transgenic mice require the availability of well characterized transcriptional regulatory elements, promoters or enhancers, that direct transgene expression in specific cell types. For example, Surfactant protein C regulatory elements have been used extensively to drive high level expression of transgenes in the lung [1,2,3,4]. Similarly, Transthyretin regulatory elements confer expression of transgenes in the liver [5, 6]. Unfortunately, the number well-characterized enhancers/promoters available for transgene expression is limited. This is due to the fact that, although in situ hybridization and RNA blotting techniques can rapidly characterize the expression patterns of genes, the identification of regulatory elements that control cell type-specific expression in animals is extremely laborious. An additional complication associated with traditional transgenic approaches is that the number and genomic location of transgene integration events is random. This can have a significant impact on both the level and site of transgene expression. These problems can be circumvented, however, by utilizing endogenous regulatory elements. This can be achieved by introducing transgenes into defined genomic loci through homologous recombination in ES cells ("knock-in") [7]. While this approach requires knowledge of the gene expression pattern it does not require any analysis of gene regulatory elements. Such a "knock-in" approach would, in effect, significantly increase the repertoire of regulatory elements available to drive transgene expression. Moreover, it ensures that a single copy of the transgene is introduced into a defined genomic location. The introduction of LacZ into specific genes during the generation of knockout mice has demonstrated that such an approach can be successful [8, 9]. The downside, however, is that every individual transgene an investigator wishes to express must be independently targeted to the chosen genomic locus. This can be tedious, especially if the rate of recombination at the chosen genomic locus is low. Here, we describe a simple method that allows transgenes targeted to a marked genomic locus to be selected with extremely high efficiency in ES cells. This approach is also appealing because it relies on reconstitution of active neomycin phosphotransferase activity, and so correctly targeted ES cell clones can be simply selected by growth in G418.
Results
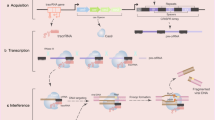
Fig 1 illustrates the overall strategy that we have used to introduce single copy transgenes into a defined genomic locus. For convenience the procedure can be considered in three distinct steps. First a specific genomic locus is chosen based on known gene expression data. In the example presented here we chose the Hnf3α locus because Hnf3α is expressed throughout the embryonic gut endoderm of the mouse and our ultimate objective is to assess the function of transgenes expressed in this tissue [10,11,12,13]. In addition, mice lacking a single Hnf3α allele have no observable phenotype, so that introduction of transgenes into this locus will not inherently affect embryonic development [14, 15]. Once the locus is chosen a targeting vector is designed to introduce a pgk/loxP-Neo cassette that confers resistance to G418 in mammalian cells. Importantly, the pgk promoter of this cassette is flanked by loxP elements so that the promoter can easily be removed using Cre recombinase. In step 2 a Cre-expression plasmid is transiently introduced into the pgk/loxP-Neo targeted ES cells. Cre-mediated deletion of the pgk promoter results in the cells reverting to G418 sensitivity while retaining the Neo gene. Step 2 is rapid and straightforward because Cre-mediated recombination is extremely efficient. The generation of these G418-sensitive ES cells containing Neo integrated into the Hnf3α locus form the basis for targeting transgenes to this locus.
Strategy used to introduce transgenes into a defined genomic locus in ES cells. For convenience the approach can be considered in three steps (boxes). In step1, a cassette containing neomycin phosphotransferase coding sequence (Neo, green) that is expressed from a phosphoglycerate kinase-1 (pgk) promoter (light blue) flanked by loxP elements (red) is targeted to a chosen locus, in this case Hnf3α (yellow), by homologous recombination. Cells containing the Neo cassette are resistant to the pharmacological inhibitor G418. In step 2, sensitivity of the targeted ES cells to G418 is restored by removing the Pgk promoter by transiently introducing a plasmid that expresses Cre recombinase. In step 3, transgenes (dark blue) are introduced to the locus through homologous recombination. The short arm of homology contains a truncated Neo gene (ΔNeo) that lacks phosphotransferase activity that can be expressed from a Pgk promoter. The long arm of homology consists of genomic DNA sequences lying 5' to the intended site of integration (not drawn to scale). Homologous recombination reconstitutes expression of Neo and generates G418 resistant ES cells. The site of transcriptional initiation of Hnf3α is indicted as a black dot with an arrow.
In step 3, a general targeting vector is constructed that will allow the introduction of transgenes to the Hnf3α/Neo locus. The salient features of such a vector are shown in Fig 2. A unique SalI restriction endonuclease site allows the introduction of coding sequences into this vector. A 3' splice site and polyadenylation signal has been incorporated to facilitate proper post-transciptional processing of transgenes. Targeting of transgenes to the Hnf3α/Neo locus is mediated by two arms of homology. The 3' arm of homology contains nucleotides 29 to 626 of the Neo coding sequence with an accompanying pgk promoter flanked by loxP elements. No G418 resistant colonies were obtained when plasmids containing this truncated Neo cassette were introduced into wild type ES cells by electroporation (data not shown). This confirmed that deletion of the 3'-end of Neo results in loss of resistance to G418 [16]. The 5' region of homology consisted of 4.5 kb of Hnf3α genomic sequence that extends into sequences encoding the untranslated 5' leader region of Hnf3α mRNA [17]. These sequences ensure that the site of transcriptional initiation remains intact after transgenes have been targeted to the Hnf3α locus [17].
Schematic of a general targeting vector used to introduce transgenes into the Hnf3α locus by homologous recombination. The 5' arm of homology contains Hnf3α genomic DNA that extends 3' of the Hnf3α transcriptional start-site (arrow). Coding sequences can be introduced into a unique Sal1 site that lies 5' to sequences containing an intron and polyadenylation signal for efficient RNA processing. A truncated Neo gene (ΔNeo) that lacks phosphotransferase activity provides the short arm of homology. Expression of ΔNeo is regulated by a Pgk promoter flanked by loxP elements. For electroporation into ES cells the targeting vector can be made linear by digesting with Sst1.
To ascertain whether the strategy proposed in Fig 1 was feasible and to determine the efficiency of such an approach we attempted to introduce a lacZ transgene into the Hnf3α locus. The genotype of ES cell clones from each step of the procedure was determined by Southern blot analysis of genomic DNA (Fig 3). Figs 3a and 3b show that the genotype of ES cells at each stage of the procedure could be distinguished by genomic Southern blot analysis using a single DNA probe (probe A) to identify a Hind III/EcoRV restriction endonuclease fragment that was specific to each targeted allele. In step1 we introduced a pgkloxP-Neo cassette into the Hnf3α locus following the procedure described in materials and methods. Fig 3b shows that probe A identifies an 8.0 kb Hind III/EcoRV fragment in wild type ES cells. Targeting of a pgkloxP-Neo cassette to this locus introduces a unique EcoRV site that generates a 4.0 kb HindIII/EcoRV fragment also detected by probe A. We obtained four correctly targeted ES cell clones out of 200 that were resistant to both G418 and gancyclovir (Fig 3b). In addition to the data shown in Fig 3b, the genotype of these colonies was confirmed by Southern blot using DNA fragments that corresponded to 3' Hnf3α genomic sequences and to Neo (not shown). These correctly targeted ES cell clones were called Hnf3αloxPNeo clones 1-4 to indicate that a pgkloxP-Neo cassette had been introduced into the Hnf3α locus. Next, a plasmid that expresses Cre-recombinase in mammalian cells was introduced into Hnf3αloxPNeo ES cells by transient transfection (materials and methods). The expression of Cre recombinase mediated recombination between the loxP elements that flanked the pgk promoter. Southern blot analysis (fig 3) shows that deletion of the pgk promoter in Hnf3αloxPNeo cells generated a predicted 3.5 kb HindIII/EcoRV fragment that was detected by probe A. Moreover, Fig 3c shows that deletion of the pgk promoter resulted in the Hnf3αloxPNeo cells reverting to G418-sensitivity because the promoter is necessary for Neo expression. The pgk promoter was deleted from 25% of transfected Hnf3αloxPNeo ES cells, using the approach described in materials and methods. ES cell lines from which the pgk promoter had been deleted were named Hnf3αΔpgk-Neo to indicate this event.
Genotype and G418 resistance of ES cells harboring insertions at the Hnf3α locus. a) A schematic showing the relative position of HindIII and EcoRV restriction endonuclease cut sites at the Hnf3α locus in ES cells. The predicted endonuclease fragment sizes identified by probe A (line) are shown above. Hnf3α ES cells have a wild type Hnf3α locus (Hnf3α, wt) and Hnf3αloxPNeo ES cells have a PgkloxP-Neo cassette introduced into a single Hnf3α allele. The pgk promoter was deleted from Hnf3αloxPNeo ES cells using Cre recombinase to generate G418 sensitive Hnf3αΔpgk-Neo ES cells. Hnf3αLacZ ES cells were generated by the introduction of a LacZ transgene into the Hnf3αΔpgk-Neo locus of Hnf3αΔpgk-Neo ES cells by homologous recombination. b) Southern blot of HindIII/EcoRV-digested genomic DNA isolated from Hnf3α, Hnf3αloxPNeo, Hnf3αΔpgk-Neo, and Hnf3α LacZ ES cells. Probe A identified an 8.0 kb wild type fragment in Hnf3α ES cells (lane 1). In Hnf3αloxPNeo (lane 2), Hnf3αΔpgk-Neo (lane 3), and Hnf3α LacZ (lane 4) ES cells probe A hybridized to an additional fragment of 4.0, 3.5 and 2.7 kb, respectively, due to the introduction of a novel EcoRV restriction endonuclease cut site. c) Hnf3α, Hnf3αloxPNeo, Hnf3αΔpgk-Neo, and Hnf3α LacZ ES cells were cultured in the absence (-G418) or presence (+G418) of 300 μg/ml of G418. Staining with methyl green identified G418-resistant ES cell colonies (blue dots).
Our ultimate goal is to target a number of different transgenes to the Hnf3α locus with the aim of expressing them ectopically throughout the embryonic gut endoderm. To facilitate this we have designed a general targeting vector that can accommodate and allow expression of any open reading frame when targeted to the Hnf3α locus in Hnf3αΔpgk-Neo ES cells. There are a number of important features associated with this vector that are illustrated in Fig 2. This plasmid includes a unique SalI restriction endonuclease cut-site for insertion of open reading frames, followed by intron and poly- [A] addition sequences for correct post-transcriptional processing of transgenic RNA. The 5' arm of homology within the targeting vector defines where the transgene is positioned relative to the Hnf3α locus. We maintained the integrity of the endogenous Hnf3α transcriptional start-site to ensure bona-fide expression of any inserted transgene. This was achieved by using a 5' arm of homology whose 3' end lay within genomic sequences encoding the untranslated leader of the Hnf3α mRNA (Fig 2) [17]. The 3' arm of homology consisted of the 5' end of Neo coding sequence accompanied by the pgk promoter with flanking loxP elements. This Neo fragment lacked sequences encoding the last 49 amino acids of neomycin phosphotransferase. Such truncation of the Neo gene disrupted its ability to encode resistance to G418 confirming the results of Beck et al (data not shown) [16]. Therefore, ES cells that randomly integrate this targeting vector would remain sensitive to G418. In contrast, we predicted that homologous recombination between the truncated Neo sequences in the targeting vector and Neo sequences within Hnf3αΔpgk-Neo ES cells would reconstitute expression of active neomycin phosphotransferase and so confer resistance to G418. This in turn implies that, following electroporation of this targeting vector into Hnf3αΔpgk-Neo ES cells, 100% of G418 resistant colonies would have undergone homologous recombination. To test this prediction we generated a targeting vector that contained the LacZ gene from E.coli. Homologous recombination should place expression of this transgene under the control of Hnf3α transcriptional regulatory elements. This plasmid was introduced into Hnf3αΔpgk-Neo ES cells by electroporation and fifteen colonies were collected that were resistant to G418. The genotype of the colonies was again ascertained by Southern blot analysis of genomic DNA. Fig 3α shows that introduction of the LacZ transgene into the Hnf3α locus was predicted to generate a 2.7 kb HindIII/EcoRV fragment that interacts with probe A. Fig 4 shows that fifteen out of fifteen ES cell lines that were resistant to G418 had undergone homologous recombination and contained the LacZ transgene correctly targeted to the Hnf3α locus. These data confirm the extraordinary efficiency of using this approach to target transgenes to a marked genomic locus.
Targeting of a LacZ transgene to the Hnf3αΔNeo locus. a) A targeting vector (materials and methods) containing a LacZ transgene was introduced into Hnf3αΔpgk-Neo ES cells by electroporation and cells were cultured in media containing G418. The genotype of ES cell clones that were resistant to G418 was determined by Southern blot analysis of genomic DNA digested with HindIII and EcoRV. A wild type 8.0 kb Hnf3α fragment and a 3.5 kb " pgk-Neo targeted" fragment from Hnf3αΔpgk-Neo ES cells (lane 1) was identified using probe A. The 3.5 kb " pgk-Neo targeted" fragment was replaced with a 2.7 kb EcoRV fragment in all fifteen G418 resistant clones examined due to the introduction of a novel EcoRV site that lies within the LacZ transgene (Hnf3αLacZ, lanes 2-16). b) Embryos at 10.5 days of gestation that were derived from Hnf3αLacZ ES cells express β-galactosidase (blue) throughout the developing hindgut (hg), foregut (fg), liver (l) and stomach (s).
We predicted that transgenes inserted into the Hnf3α locus would be expressed throughout the endoderm of the developing gut. To determine if this was true we generated embryos from ES cells containing LacZ at the Hnf3α locus (Hnf3αLacZ ES cells) and stained for expression of β-galactosidase. Mouse embryos derived solely from ES cells were generated by tetraploid aggregation as described previously [18,19,20]. A total of 10 embryos were produced and all showed the same pattern of β-galactosidase expression. Fig 4b shows β-galactosidase staining in Hnf3αLacZ embryos at 10.5 days of embryonic development. As expected, β-galactosidase was expressed throughout the gut, liver and at particularly high levels in the developing stomach. Endogenous Hnf3α is also expressed in the floorplate of the neural tube as well as the notochord. However, expression of β-galactosidase in Hnf3αLacZ transgenic embryos was undetectable in these tissues. In generating the Hnf3αLacZ targeting vector we deleted all genomic sequences lying 3' to the first intron of the Hnf3α gene. It is likely that this intron or other untranslated sequences contain regulatory elements that specifically direct expression of Hnf3αLacZ in the floorplate of the neural tube and notochord [21]. Indeed when LacZ is introduced into exon 2 in ES cells, leaving the first intron intact, expression of β-galactosidase is readily detectable in the neural-tube floorplate and notochord [9].
Discussion and conclusions
We have described a method that allows transgenes to be easily targeted to a defined locus in the mouse genome in ES cells. Although this is a multiple-step procedure, once the chosen locus has been targeted using homologous recombination, subsequent manipulations are extremely efficient. Indeed in the final step, where the transgene of interest is targeted to the desired locus, we found that 100% of G418 resistant ES cell colonies were correctly targeted. Transgenic mice and embryos can be generated from these ES cells by standard injection into blastocysts and subsequent breeding of the resulting chimeric mice. It is worth noting, however, that if germline transmission is the aim of the experiment it is important to ensure that the "Δpgk-Neo" cells generated in step 2 are germline competent. This is important because each round of clonal selection increases the likelihood of losing germline competency of the ES cells.
The targeting of transgenes into a given locus has a number of advantages over traditional methods of generating transgenic mice. Traditionally, transgenic mice are produced by injection of DNA into the male pronucleus of fertilized mouse eggs. A variable number of copies of the transgene are then integrated into the mouse genome at random locations. The position and number of transgene copies can have a profound effect on their expression. In some cases expression is repressed or, in contrast, undesirably activated at ectopic sites. When transgenes are targeted to a known genomic locus such variation is avoided and expression of the transgene is significantly more predictable [7]. Such control over the site and level of expression is important because variations could have unpredictable impacts on the phenotype presented by the transgenic mice.
Introduction of single copy transgenes into the hydroxyphosphoribosyl transferase (HPRT) locus has been previously described by Bronson et al. This approach successfully overcomes the problems associated with the integration of variable copy numbers of transgenes into random genomic locations. However, it is only suitable for introduction of transgenes to the HPRT locus and, in addition, requires the availability of HPRT-negative ES cell lines. Cre-mediated targeting of single copy transgenes to specific genomic sites that have previously been marked by loxP elements has also been described both in somatic cells and more recently in ES cells using a double LoxP targeting strategy [22, 23]. Although this approach also results in efficient targeting to a defined genomic locus it relies on co-transfection of a Cre expression plasmid along with a loxP-targeting vector that carries the transgene. Hardouin et al also recently described an elegant approach to introduce transgenes to loci identified by gene trapping. Here integration of the transgene at the gene-trap locus was again mediated by Cre recombinase and translation of the transgene was facilitated by an internal ribosomal entry site (IRES) [24]. In contrast, our approach relies simply on the reconstitution of resistance to G418 and can be used to target transgenes to any genomic locus.
Transgenic mice are often used to examine gene function through ectopic expression studies. This requires the availability of characterized transcriptional regulatory elements that are capable of expressing transgenes in the tissue of choice. In addition, for developmental studies, the transcriptional regulatory elements have to ensure transgene expression during the correct developmental time frame. Although the expression patterns of many genes have been described in detail, characterization of promoter and enhancer elements that control this expression is much more limited. This is partly due to the fact that complex expression patterns often utilize genomic regulatory sequences that are positioned many kilobases away from the gene making them difficult to identify. However, the introduction of transgenes into specific loci allows the utilization of intact endogenous transcription regulatory sequences, which increases the likelihood that the transgene will be expressed in the expected fashion.
Using targeted ES cells also provides the potential of expressing lethal transgenes. This may be important for examining the effects of expressing gain-of-function or dominant-negative alleles of a gene product. This is difficult using a conventional approach because of the need to establish founder mice expressing the transgene. However, if a line of "transgenic" ES cells can be established then it is possible to generate clonal embryos directly from these ES cells by aggregating them with tetraploid embryos. Indeed, here we have used this approach to establish that the introduction of a LacZ transgene into the Hnf3α locus facilitates transgene expression throughout the developing gut (Fig 4).
In sum, we have described a method that facilitates the efficient introduction of transgenes into predefined loci in the mouse genome. By selecting appropriate sequences for homologous recombination this approach can be tailored toward any specific genomic locus. We, therefore, believe that this approach expands the repertoire of tools available for genetic manipulation in the mouse and will enhance our ability to address gene function in mammals.
Materials and Methods
Plasmids
In the following cloning steps "blunt" infers that the cohesive ends of a DNA fragment, cut by restriction endonucleases, were repaired by the Klenow fragment of DNA PolI in the presence of deoxyribonucleotides.
Hnf3α targeting vector(p3αloxPNeo-TK)
A 900 bp Xho1/Xba1 fragment of Hnf3α genomic DNA that included exon 1 and a portion of intron 1 was used as the 5' arm of homology. The 3' arm of homology was cloned as a 4.5 kb Xho1 fragment of Hnf3α genomic DNA. A Xho1/HindIII (blunt) cassette containing the Tn5 neomycin phosphotransferase (Neo)gene, that could confer resistance to G418 and whose expression in mammalian cells was directed by a phosphoglycerate kinase-1 (pgk) promoter, was introduced into an XbaI (blunt) site between the Hnf3α genomic sequences [16, 25]. The pgk promoter was flanked by loxP elements so that it could be deleted by the action of Cre recombinase. The herpes simplex virus thymidine kinase (hsv-TK) gene, whose expression was also regulated by the pgk promoter, was introduced adjacent to the Hnf3α 5' arm of homology to provide negative selection in the presence of gancylovir.
Transgene targeting vector (p3αΔNeo)
The transgene-targeting vector used in these experiments was generated in multiple steps (Fig 2). A NotI/RsrII (blunt)1.4 kb cassette containing the 5' end of Neo coding sequence accompanied by the pgk promoter with flanking loxP elements was introduced into the SphI site (blunt) of pNEB193 (New England Biolabs). Deletion of the 49 c-terminal codons of the Neo gene disrupted its ability to encode resistance to G418 (data not shown) [16]. This sub-fragment of Neo provided the 3' arm of homology in the targeting vector. A 530 bp EcoRI fragment (blunt) containing an intron from the mouse protamine gene as well as poly(A) addition sequences was isolated from the plasmid pLacF and cloned into the PmeI site (blunt) of the preceding plasmid [26]. Finally the 5' arm of homology was provided by a 4.5 kb Xho I fragment of Hnf3α genomic DNA that was introduced into a unique Pac1 site (blunt) in the preceding plasmid. Importantly, this fragment extended from 5' genomic DNA into sequences encoding the untranslated 5' end of Hnf3α mRNA. This cloning strategy left a unique Sal I site into which coding sequences of transgenes could be introduced. A NotI (blunt) 3.6 kb lacZ fragment from pCMV-βgal (Clontech) was ligated into this SalI (blunt) site to generate a targeting vector that could be used to introduce lacZ into the "marked" Hnf3α locus.
Culture and selection of embryonic stem cell lines
All ES cell lines were cultured on mitotically inactivated primary embryonic fibroblasts in ES cell medium supplemented with recombinant leukemia inhibitory factor (LIF) as described elsewhere [27]. Gene targeting was carried out using 100 μg of linear targeting plasmid. This was introduced into 2.5 × 108 ES cells by electroporation at 250 volts/500μf/resistance 8 using a BTX ECM600 electroporation system. Cells were plated on thirteen 10 cm2 tissue culture dishes and grown for two days in ES cell medium supplemented with LIF. Cells containing Neo were selected by supplementing the ES cell medium with 300 μg/ml Geneticin (G418,Gibco-BRL) and negative selection against Hsv-tk gene expression was achieved by including 2 μM gancyclovir (Roche). Recombination between loxP elements in ES cells was mediated by introducing a Cre expression plasmid, pHDMCCre8 (provided by Dr. Klaus Kaestner). 100 μg of pHDMCCre8 was introduced into 5 × 107 ES cells by electroporation using 400 v/500μf/resistance setting 8. 1/100,000 of the total electroporated cell population was plated per 10 cm2 tissue culture dish in ES cell medium supplemented with LIF and grown until individual colonies could be collected.
Tetraploid aggregation and β-galactosidase staining
Embryos were generated from ES cells by aggregating them with 4-cell stage embryos made tetraploid by electrofusion, as described previously [19, 20, 27]. Aggregates that formed blastocysts after overnight culture were allowed to continue their development in utero by transferring them to a pseudopregnant surrogate mother. Embryos were stained for expression of β-galactosidase using standard techniques [28].
Abbreviations
- Neo:
-
neomycin phosphotransferase
- pgk:
-
phosphoglycerate kinase promoter
- Hsv-TK:
-
Herpes simplex virus thymidine kinase
- Hnf:
-
hepatocyte nuclear factor
- LIF:
-
leukemia inhibitory factor
- HPRT:
-
hydroxyphosphoribosyl transferase
References
Zeng X, Wert SE, Federici R, Peters KG, Whitsett JA: VEGF enhances pulmonary vasculogenesis and disrupts lung morphogenesis in vivo. Dev Dyn. 1998, 211: 215-227. 10.1002/(SICI)1097-0177(199803)211:3<215::AID-AJA3>3.0.CO;2-K.
Geraci MW, Gao B, Shepherd DC, Moore MD, Westcott JY, Fagan KA, Alger LA, Tuder RM, Voelkel NF: Pulmonary prostacyclin synthase overexpression in transgenic mice protects against development of hypoxic pulmonary hypertension. J Clin Invest. 1999, 103: 1509-1515.
Hoyle GW, Li J, Finkelstein JB, Eisenberg T, Liu JY, Lasky JA, Athas G, Morris GF, Brody AR: Emphysematous lesions, inflammation, and fibrosis in the lungs of transgenic mice overexpressing platelet-derived growth factor. Am J Pathol. 1999, 154: 1763-1775.
Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL: Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999, 126: 4005-4015.
Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T: Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996, 10: 245-260.
Rausa FM, Tan Y, Zhou H, Yoo KW, Stolz DB, Watkins SC, Franks RR, Unterman TG, Costa RH: Elevated levels of hepatocyte nuclear factor 3beta in mouse hepatocytes influence expression of genes involved in bile acid and glucose homeostasis [In Process Citation]. Mol Cell Biol. 2000, 20: 8264-8282. 10.1128/MCB.20.21.8264-8282.2000.
Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O: Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci U S A. 1996, 93: 9067-9072. 10.1073/pnas.93.17.9067.
Weinstein DC, Altaba ARi, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE: The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell. 1994, 78: 575-588.
Duncan SA, Navas AM, Dufort D, Rossant J, Stoffel M: Regulation of a transcription factor network required for cell differentiation and metabolism. Science. 1998, 281: 692-695. 10.1126/science.281.5377.692.
Ang S-L, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS: The formation and maintainance of the definitive endoderm lineage in the mouse: involvement of HNF-3/ forkhead proteins. Development. 1993, 119: 1301-1315.
Altaba ARi, Prezioso VR, Darnell JE, Jessell TM: Sequential expression of HNF3β and HNF3α by embryonic organizing centers: the dorsal lip/node, notochord and floor plate. Mech. Dev. 1993, 44: 91-108. 10.1016/0925-4773(93)90060-B.
Sasaki H, Hogan BLM: Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouseembryo. Development. 1993, 118: 47-59.
Monaghan AP, Kaestner KH, Grau E, Schutz G: Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3α, β, and γ genes in determination of the difinitive endoderm, chordamesoderm, and neurectoderm. Development. 1993, 119: 567-578.
Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G: Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999, 13: 495-504.
Shih DQ, Navas MA, Kuwajima S, Duncan SA, Stoffel M: Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3alpha-deficient mice. Proc Natl Acad Sci U S A. 1999, 96: 10152-10157. 10.1073/pnas.96.18.10152.
Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H: Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982, 19: 327-336. 10.1016/0378-1119(82)90023-3.
Kaestner KH, Hiemisch H, Lucknow B, Schutz G: The HNF-3 gene family of transcription factors in mice: gene structure,cDNA sequence, and mRNA distribution. Genomics. 1994, 20: 377-385. 10.1006/geno.1994.1191.
Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J: Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990, 110: 815-821.
Nagy A, Rossant J: Production of completely ES cell-derived fetuses. In: Gene targeting: A practical approach Edited by Joyner A. pp. 147-179. Oxford, UK: Oxford Unviersity Press;. 1993, 147-179.
Duncan SA, Nagy A, Chan W: Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of HNF-4-/- embryos. Development. 1997, 124: 279-287.
Clevidence DE, Zhou H, Lau LF, Costa RH: The -4 kilobase promoter region of the winged helix transcription factor HNF-3alpha gene elicits transgene expression in mouse embryonic hepatic and intestinal diverticula. Int J Dev Biol. 1998, 42: 741-746.
Fukushige S, Sauer B: Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci U S A. 1992, 89: 7905-7909.
Soukharev S, Miller JL, Sauer B: Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res. 1999, 27: e21-10.1093/nar/27.18.e21.
Hardouin N, Nagy A: Gene-trap-based target site for cre-mediated transgenic insertion. Genesis. 2000, 26: 245-252. 10.1002/(SICI)1526-968X(200004)26:4<245::AID-GENE50>3.0.CO;2-9.
Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC: Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991, 65: 1153-1163.
Peschon JJ, Behringer RR, Brinster RL, Palmiter RD: Spermatid-specific expression of protamine 1 in transgenic mice. Proc Natl Acad Sci U S A. 1987, 84: 5316-5319.
Joyner AL: Gene Targeting: A practical approach. Oxford: Oxford University Press;. 1993
Hogan B, Beddington R, Constantini F, Lacy E: Manipulating the mouse embryo: a laboratory manual, second edn. New York: Cold Spring Harbour Laboratory Press;. 1994
Acknowledgements
We would like to thank Dr. Paula Traktman for critical reading of the manuscript and Dr. Klaus Kaestner for generously supplying the Cre expression vector and for useful advice. This work was supported by NIH R01 DK55743 to S.A.D.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhao, R., Fahs, S.A., Weiler, H. et al. An efficient method to successively introduce transgenes into a given genomic locus in the mouse. BMC Dev Biol 1, 10 (2001). https://doi.org/10.1186/1471-213X-1-10
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-213X-1-10