Abstract
Background
Escherichia coli DNA topoisomerase I binds three Zn(II) with three tetracysteine motifs which, together with the 14 kDa C-terminal region, form a 30 kDa DNA binding domain (ZD domain). The 67 kDa N-terminal domain (Top67) has the active site tyrosine for DNA cleavage but cannot relax negatively supercoiled DNA. We analyzed the role of the ZD domain in the enzyme mechanism.
Results
Addition of purified ZD domain to Top67 partially restored the relaxation activity, demonstrating that covalent linkage between the two domains is not necessary for removal of negative supercoils from DNA. The two domains had similar affinities to ssDNA. However, only Top67 could bind dsDNA with high affinity. DNA cleavage assays showed that the Top67 had the same sequence and structure selectivity for DNA cleavage as the intact enzyme. DNA rejoining also did not require the presence of the ZD domain.
Conclusions
We propose that during relaxation of negatively supercoiled DNA, Top67 by itself can position the active site tyrosine near the junction of double-stranded and single-stranded DNA for cleavage. However, the interaction of the ZD domain with the passing single-strand of DNA, coupled with enzyme conformational change, is needed for removal of negative supercoils.
Similar content being viewed by others
Background
Escherichia coli DNA topoisomerase I is a representative example of type IA DNA topoisomerase (for reviews, see refs [1, 2]). Its major biological role in the bacterial cell is the removal of excessive negative supercoils from DNA to maintain the DNA at optimal superhelical density along with DNA gyrase [3]. The enzyme has a molecular weight of 97 kDa and the active site tyrosine responsible for DNA cleavage is found in the 67 kDa N-terminal transesterification domain. The structure of this 67 kDa domain has been determined by X-ray crystallography to be torus-like, indicating the need for protein conformational change for strand passage to take place after DNA cleavage [4]. Relaxation activity requires the presence of the Zn(II) binding tetracysteine motifs [5] found between the 67 kDa N-terminal domain (Top67) and the 14 kDa C-terminal single-stranded DNA binding domain (Figure 1). The three tetracysteine motifs do not form a stably folded structure on its own, but when combined with the 14 kDa C-terminal domain, forms a stably folded 268 amino acid DNA binding domain (ZD domain) that has higher affinity for single-stranded DNA than the 121 amino acid 14 kDa C-terminal region by itself [6]. Recent sequence and structural analysis suggests that the 14 kDa domain is evolutionarily related to the three tetracysteine motifs and belongs to the zinc ribbon family [7]. The ZD domain in E. coli topoisomerase I probably evolved from a domain that binds five Zn(II) originally.
Removal of negative supercoils from DNA by bacterial type IA topoisomerase involves the following steps: (1) binding of the enzyme to the junction of double-stranded and single-stranded DNA [8]; (2) cleavage of a single-strand of DNA near the junction with cleavage sequence preference of a cytosine in the -4 position to form the covalent intermediate [9, 10]; (3) conformational change of the covalent enzyme-DNA complex to result in physical separation of the 5' phosphate covalently linked to the active tyrosine, and the 3' hydroxyl of the cleaved DNA; (4) passage of the complementary single strand through the break; (5) enzyme conformational change to bring the 5' phosphoryl end back into the proximity of the 3' hydroxyl group of the cleaved DNA; (6) religation of the phosphodiester bond. Although it is known that the ZD domain can function as a DNA binding domain, its exact role in these individual steps of removal of a negative superhelical turn from DNA by E. coli topoisomerase I remains to be defined. Using purified 67 kDa transesterification domain and 30 kDa ZD domain, results from experiments described here provide new insight into the action of these two individual domains in the enzyme mechanism.
Results
Partial restoration of relaxation activity from mixing of Top67 and ZD domains
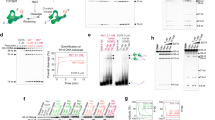
As reported previously [11], the N-terminal transesterification domain Top67 by itself did not exhibit any relaxation activity when assayed with negatively supercoiled plasmid DNA (Figure 2a). The 30 kD C-terminal ZD domain also had no relaxation activity by itself, as expected. Partial relaxation of the input supercoiled DNA was detected when Top67 was mixed with the ZD domain prior to addition of DNA. A ratio of 2 ZD molecules added for each Top67 was found to be sufficient for maximum relaxation activity, with no increase in activity when higher ratio of ZD/Top67 was used (data not shown). The specific activity observed under this optimized condition (Figure 2a) was still about 10 fold lower than that of the intact enzyme. Analysis of the time course of relaxation with 6 pmoles of topoisomerase I or top67 reconstituted with ZD (Figure 2b) showed that negative supercoils were removed at a much slower rate by the reconstituted activity.
Partial restoration of relaxation activity by complementation of Top67 and ZD domains. (a). Agarose gel electrophoresis was carried out to analyze the relaxation reaction products after 1 h of incubation. Lane 1: supercoiled plasmid DNA with no protein added; lane 2: Top67 alone; lane 3: ZD alone. Lanes 4–7 (Top67 reconstituted with ZD) and lanes 8–11 (topoisomerase I) have 6, 1.2, 0.24 and 0.05 pmoles of proteins added. (b). Time course of relaxation reaction catalyzed by 6 pmoles of topoisomerase I, or Top67 reconstituted with ZD
Top67 and ZD domains have comparable binding affinities to single-stranded DNA but significantly different affinities for double-stranded DNA
The gel mobility shift assay was used to compare the binding affinities of Top67 and the ZD domain to a 5' end-labeled single-stranded oligonucleotide 35 base in length. As shown in Figure 3a, these two domains had similar affinities for binding to the single-stranded substrate. The half maximal binding values based on the average of results from three different experiments were 0.02 μM for Top67 and 0.04 μM for the ZD domain. However, with the same oligonucleotide in a duplex form (Figure 3b), Top67 exhibited much higher affinity (half maximal binding value = 0.07 μM) than the ZD domain (half maximal binding value > 5 μM).
Binding of Top67 and ZD to single-stranded and double-stranded DNA. The gel mobility shift assay was used to compare the binding affinities. The substrates used are (a): single-stranded 5'GAAAACTCACAGGAAGCGGCCGAAGCGATTCGTCC 3'; (b): the same labeled strand of hybridized to its complementary strand. Open circles: Top67; solid circles: ZD.
Top67 can recognize cleavage sites preferred by E. coli DNA topoisomerase I
Previous studies have shown that E. coli DNA topoisomerase I cleavage of single-stranded DNA occurs with selectivity for sites with the C nucleotide base at the – 4 position [9, 10] and that the enzyme preferentially cleaves at junctions of double-stranded and single-stranded DNA [8]. Several different 5'-end labeled substrates were prepared and used in cleavage assays to compare the cleavage sites selected by Top67 versus topoisomerase I. The results showed that with single-stranded substrates, Top67 also preferred cleavage sites with a C nucleotide base at the -4 position as reported for most of the type IA topoisomerases [12]. There were some differences from topoisomerase I in the relative distribution of cleavage products among the potential cleavage sites (Figure 4a,4b). Top67 appeared to be more non-discriminatory in selection of the possible cleavage sites with the C nucleotide in the -4 position. Addition of the ZD domain had no effect on the cleavage selectivity of Top67. A substrate with both single-stranded and double-stranded regions was constructed to mimic such junction in negatively supercoiled DNA. Top67 and topoisomerase I recognised the same cleavage site on this substrate (Figure 4c). Maximal yield of cleavage products was obtained for both Top67 and topoisomerase I within seconds after mixing of the enzyme and DNA so any potential difference in cleavage rates between the Top67 and topoisomerase I is unlikely to account for the difference in relaxation efficiency.
Cleavage selectivity of topoisomerase I and Top67. This was analyzed using single-stranded 32mer (a), 31mer (b), or substrate with both single- and double-stranded regions (c). For (a) and (b), lane 1: no protein added, lane 2: topoisomerase I, lane 3: Top67, lane 4: Top67 mixed with ZD. For (a), lane 5: ZD alone. For (c), lane 1: topoiosmerase I, lane 2: Top67, lane 3: no protein added, lane 4: DNase I digestion pattern.
Top67 cleavage sites are religated upon addition of high salt and Mg2+
To test the religation capability of Top67, a 5'-end labeled oligonucleotide 61 base in length was first incubated with the enzyme in low ionic strength buffer to allow formation of the cleaved complex. Sodium chloride concentration was then increased to 1 M to induce reversal of cleavage and dissociation of the enzyme from the DNA. We observed that more complete and consistent reversal of cleavage was obtained with both topoisomerase I and Top67 if a low concentration of Mg2+ (4 mM) was also added with the NaCl. This is consistent with an early observation of dissociation of the enzyme-DNA complex in high salt upon addition of Mg2+[13]. It has also been reported [14] that addition of Mg2+ was apparently not required for observation of this reversal of cleavage. However, it is possible that some enzyme preparations may contain bound Mg2+ and the low concentration of bound Mg2+ might have been sufficient for reversal of cleavage, as postulated previously to explain the data [14]. The results of this cleavage reversal experiment (Figure 5) indicated that the ZD domain was not required for efficient reversal of cleavage and Top67 could carry out religation of cleaved DNA. Again the reversal of cleavage was complete for both Top67 and topoisomerase I within seconds after the addition of high salt and Mg2+ even when the reactions were carried out on ice (data not shown) so the lack of relaxation activity by Top67 is unlikely to be due to deficiency in religation.
Reversal of DNA cleavage by topoisomerase I and Top67. A 5'-end labeled 61mer was used as substrate. C: no enzyme added. Lane 1: enzyme cleavage reaction stopped with SDS; Lane 2: enzyme cleavage reaction incubated with 1 M NaCl before SDS treatment; Lane 3: enzyme cleavage reaction incubated with 1 M NaCl and 4 mM MgCl2 before SDS treatment.
The ZD domain is not required for catenation of double-stranded DNA circles
E. coli topoisomerase I can catalyze catenation of double-stranded DNA circles if the molecules contain single-strand scissions [15, 16]. To test if the Top67 can carry out double-stranded DNA passage at enzyme cleavage sites across from the DNA nicks, the yield of DNA catenanes were compared with that obtained with topoisomerase I. In contrast to the relaxation activity, the catenating activity of Top67 shown in figure 6 was as efficient as that of full-length topoisomerase I, and the addition of the ZD domain had no effect (Figure 6a). The rate of catenane formation for Top67 alone was similar to that of topoisomerase I (Figure 6b). This catenation activity observed with topoisomerase I and Top67 was unlikely to be due to contaminating topoisomerase III activity since it was not observed with the ZD domain purified under almost identical procedures and a site-directed mutant with substitution of the active site Tyr319 by phenylalanine also did not exhibit this activity (Figure 6a).
Catenation of nicked double-stranded DNA circles by topoisomerase I and Top67. (a). Phage PM2 DNA circles with one or more single-strand scissions were incubated with 5 pmoles of proteins for 1 h. Lane 1: no enzyme added; Lane 2: topoisomerase I; Lane3: Top67; Lane 4: Top67 and ZD domain; Lane 5: ZD domain; Lane 6: mutant with Y319F substitution. (b). Aliquots of the reaction for topoisomerase I and Top67 were removed at different time points to analyse the time course of catenation.
Discussion
There are two homologous type IA topoisomerases present in E. coli. Topoisomerase III has potent DNA decatenating activity for resolution of plasmid DNA replication intermediates, but much weaker relaxation activity than topoisomerase I [17]. To exhibit maximal relaxation activity, topoisomerase III requires high temperature (52°C) along with low magnesium and monovalent ion [17, 18]. In contrast, E. coli topoisomerase I was not active in the in vitro assay for resolution of plasmid DNA replication intermediates [19]. Removal of the C-terminal 49 amino acids from the 653 amino acid topoisomerase III protein resulted in drastic reduction of catalytic activity [20]. Fusion of the carboxyl-terminal 312 amino acid residues of E. coli topoisomerase I, which includes the entire ZD domain, onto the 605 N-terminal amino acids of topoisomerase III generated a hybrid topoisomerase that has relaxation activity resembling topoisomerase III along with weak decatenating activity [21]. Although preferring single-stranded DNA as binding substrate, topoisomerase I had been shown to also bind double-stranded DNA [22], but there is no data available to indicate which domain in the enzyme is responsible for this interaction.
The experiments described here measured directly the interaction of the ZD domain with both double-stranded and single-stranded DNA substrates. ZD domain was found to bind to single-stranded DNA, but not double-stranded DNA, with high affinity. This result indicated that with regard to the mechanism of E. coli topoisomerase I, the ZD domain was likely to function as a single-stranded DNA binding domain instead of having double-stranded DNA binding function as previously suggested [21]. Even though Zn(II) binding transcription factors that recognise specific double-stranded DNA are well represented in eukaryotes [23, 24], there are also numerous examples of Zn(II) coordination being required for interaction with single-stranded nucleic acid or damaged DNA with single-strand characteristics [24–27].
The effect of removal of the ZD domain on the individual step of enzyme action was also investigated using Top67. The results indicated that Top67 was effective in binding to both double-stranded and single-stranded DNA. As a result, Top67 could position itself in the absence of ZD domain at the junction of double- and single-stranded DNA for subsequent DNA cleavage, as previously observed for intact topoisomerase I [8]. Reversal of DNA cleavage also took place readily with Top67 upon addition of 1 M NaCl and 4 mM MgCl2. The ZD domain also was not required for selectivity of a cytosine in the -4 position relative to the cleavage sites.
Despite its ability to recognise the DNA substrate and carry out DNA cleavage-religation, Top67 by itself cannot catalyze change of linking number in the relaxation of supercoiled DNA. The single-strand DNA substrate designated for the ZD domain in the catalytic mechanism of the enzyme may be the strand of DNA complementary to the strand first cleaved by the enzyme to form the covalent complex. This interaction with the passing strand of DNA would not be needed for the first two steps of enzyme mechanism up to the formation of the covalent complex. Our results showed that adding the purified ZD domain partially restored the relaxation activity. Therefore the ZD domain can supply the function that is missing in Top67 even when the two domains are not covalently linked. However, the resulting relaxation activity is much less efficient than that of the intact enzyme, suggesting that coordinated actions of the two domains are required for efficient removal of negative supercoils from DNA. The requirement of specific protein-protein interactions between the two domains could also account for the weak relaxation activity observed for the hybrid topoisomerase with ZD linked to topoisomerase III sequence [21].
This proposed role for the ZD domain in interacting with the passing single-strand of DNA is also supported by the observation that there is no difference between Top67 and intact topoisomerase I in the formation of catenanes. This reaction involves passage of another double-stranded DNA circle, instead of the complementary DNA strand through the break generated by DNA cleavage so the ZD domain would not be expected to play any significant role. High concentration of DNA substrate is required to favor formation of catenanes catalyzed by topoisomerase I, and the enzyme also has to be present in higher concentration compared to the relaxation reaction. The double-stranded DNA-binding activity in E. coli topoisomerase III required for highly efficient decatenation activity is attributed to a 17-amino-acid residue with no counterpart in E. coli topoisomerase I [28, 29]. It may be required for interaction with the passing double-strand of DNA in the decatenation mechanism. The presence of this decatenation loop instead of the Zn(II) binding ZD domain in topoisomerase III may account for the dominance of the decatenation activity over the relaxation activity.
Based on these results, we propose a model for the relaxation of supercoiled DNA by E. coli topoisomerase I (Figure 7) modified from previous versions that have a number of common features but differ most significantly in the role of the Zn(II) binding domain [2, 4, 21, 29, 30]. In this model, the subdomains in Top67 is responsible for interacting with the G-strand of DNA both upstream and downstream of the cleavage site. The ZD domain interacts with the passing single-strand DNA to be transported (T-strand). After cleavage of the DNA gate strand which becomes covalently linked to Tyr319 on Top67 (step 2), protein conformational change involving both Top67 and the ZD domain increases the distance between the covalently bound 5' phosphate and non-covalently bound 3' hydroxyl of the cleaved DNA gate strand while the passing DNA strand (T-strand) is guided through the "gate" via interaction with the ZD domain (step 3) to lead to change in linking number. A second enzyme conformational change positions the cleaved DNA ends for religation (step 4). The ZD domain can still interact with the T-strand of DNA even when not linked to Top67 in the same polypeptide, but efficiency of catalysis is reduced as a result, probably due to loss of coordinated action by the two domains. The presence of the ZD domain may enhance the transition of Top67 from a closed conformation to a more open conformation so that strand passage can take place through the "DNA gate". Previous data showed that although Zn(II) binding is not absolutely required for formation of the cleaved complex, it increased the amount of cleaved complex that can be isolated [31]. When linked to Top67, the ZD domain also has some influence on the cleavage site selections. It has previously been observed that a mutation in the Zn(II) binding motif can affect the cleavage site selectivity of topoisomerase I [32] even though Top67 by itself can recognize both the cytosine in the -4 position and the junction of single- and double-stranded DNA. To gain further details for this model of enzyme action, we are characterizing the protein-protein interactions between the Top67 transesterification domain and the ZD domain, as well as the protein conformational changes that can take place when the enzyme interacts with DNA substrate.
Model for removal of a negative supercoil by E. coli DNA topoisomerase I. Subdomains I, II, III, IV found in the crystal structure of Top67 (4) are illustrated schematically along with the potential site for ZD domain (Z). G-strand: DNA strand cleaved to provide "DNA Gate". T-strand: DNA strand to be transported through the "DNA Gate".
The hyperthermophilic topoisomerase I from Thermotoga maritima has been shown to coordinate one Zn(II) with a unique tetracysteine motif Cys-X-Cys-X16-Cys-X-Cys but Zn(II) binding is not required for relaxation activity [33]. The sequence of this unique tetracysteine motifs is somewhat different from those present in other type IA topoisomerases in that the other tetracysteine motifs always had at least two amino acids separating the pairs of cysteines (Cys-X2-11-Cys), instead of just one amino acid (Cys-X-Cys) in T. maritima topoisomerase I [33]. Therefore the structure and function of the single Zn(II) binding motif in T. maritima may differ from the multiple Zn(II) binding motifs in E. coli topoisomerase I. Direct interaction between DNA and the T. maritima Zn(II) binding motif has not been demonstrated. It has been suggested that the mechanisms of these two enzymes may be different [33]. Direct interaction between the enzyme and the passing strand may not be necessary for the T. maritima topoisomerase I activity. The relaxation and decatenation activities of T. maritima topoisomerase I appear to be significantly more efficient than those of the E. coli topoisomerase I [33]. Based on their primary sequences, a number of bacterial topoisomerase I enzymes do not appear to coordinate any Zn(II) with tetracysteines motifs while other type IA topoisomerase has up to 4 tetracysteine motifs [7]. The topoisomerase I from Mycobacterium smegmatis has been demonstrated biochemically not to bind Zn(II) [34]. In contrast, mutation disrupting the fourth Zn(II) motif of Helicobacter pylori topoisomerase I abolished enzyme function in vivo[35]. Therefore there may be significant differences in the mechanisms of type IA topoisomerases from different organisms with respect to requirement of Zn(II) binding for relaxation activity.
There is also another possible explanation for the varied number of tetracysteine motifs and requirement of Zn(II) for relaxation activity found in different type IA topoisomerases. The 14 kDa C-terminal region of E. coli topoisomerase I has been classified based on its structure to be in the Zn-ribbon superfamily [SCOP release 1.50, 7] even though it does not bind Zn(II). It also has high affinity for binding to single-stranded DNA on its own when separated from the three tetracysteine motifs [36]. Based on the structural and DNA-binding properties of the E. coli topoisomerase I 14 kDa domain, one can conclude that it is possible for a subdomain in topoisomerase I to lose the Zn(II) binding cysteines during evolution and still maintains the Zn-ribbon structure and single-strand DNA binding properties [7].
Finally, the in vivo catalytic activities of eukarytotic type IA topoisomerases, the topoisomerase III from various higher organisms may be related to their sequences. The transesterification domains of these eukaryotic enzymes have high degrees of identity to E. coli DNA topoisomerase III [7, 37]. However, the decatenation loop is not present in the eukaryotic topoisomerase III sequences and to date the decatenation activity has not been demonstrated for these enzymes. The number of potential Zn(II) binding cysteine motifs range from none in S. cerevisiae DNA topoisomerase III to four highly conserved tetracysteine motifs in the beta family of the topoisomerase III enzymes [38]. The Zn(II) domain formed by these tetracysteine motifs may be required for interaction with single-strand DNA in removal of hypernegative supercoils [39] or disruption of early recombination intermediates between inappropriately paired DNA molecules [40].
Conclusions
We have shown that the ZD domain of E. coli DNA topoisomerase I is not required for the substrate recognition and DNA cleavage-religation action of the enzyme. We propose that the ZD domain interacts with the passing single-strand of DNA in the relaxation of negatively supercoiled DNA by this enzyme.
Materials and methods
Enzyme and DNA
E. coli DNA topoisomerase I and the ZD domain were expressed and purified as described [6, 41]. To express the 67 kDa N-terminal transesterification domain (Top67), a stop codon at amino acid 598 was introduced into plasmid pJW312 [42] used for topoisomerase I expression by site-directed mutagenesis employing the Chameleon-Mutagenesis kit from Stratagene. Top67 was expressed and purified with the same procedures as topoisomerase I.
The oligonucleotides were custom synthesized by Genosys. The single-strand substrates and the top strand of the duplex substrates were labeled at the 5' termini with T4 polynucleotide kinase and γ32P-ATP. The labeled oligonucleotides were purified by electrophoresis in a 12 or 15% sequencing gel. After elution from the gel slice, the labeled single-stranded oligonucleotides were desalted by centrifugation through a Sephadex G10 spin column.
The duplex or heteroduplex substrates were prepared by mixing the labeled top strand with 4 fold excess of the unlabeled bottom strand, heating at 80°C for three minutes, cooling to room temperature and purified by electrophoresis in a 20% non-denaturing polyacrylamide gel with TBE buffer.
Plasmid pJW312 DNA used in relaxation assay was purified by CsCl centrifugation. Phage PM2 DNA was extracted from infected Pseudoalteromonas espejiana cells [43] and PM2 DNA with one or more single-chain scissions used in the catenation assay was prepared as described [44].
DNA relaxation assay
Top67 and the ZD domains at different concentrations were mixed and incubated at 37°C for 10 min before addition to the 0.3 μg of supercoiled plasmid DNA in 20 μl of 10 mM Tris-HCl pH 8.0, 2 mM MgCl2, 0.1 mg/ml gelatin. After incubation at 37°C for up to 1 h, the reaction was stopped by addition of 50 mM EDTA and electrophoresed in a 0.7% agarose gel and visualized by ethidium bromide staining as described [45].
Gel mobility shift assay
The proteins were mixed with the 1 pmole of the labeled DNA substrates in 10 μl of 20 mM Tris-HCl pH 8.0, 100 μg/ml BSA, 12% glycerol and 0.5 mM EDTA. The samples were incubated at 37°C for 5 min and then loaded onto a 6% polyacrylamide gel and electrophoresed with buffer of 45 mM Tris-borate pH 8.3, 1 mM EDTA. Electrophoresis was carried out at room temperature at 2 V/cm for 2 h. After drying of the gel, bands corresponding to the protein-bound oligonucleotides and unbound oligonucleotides were visualized by autoradiography, excised and counted in a Scintillation counter for quantitation.
DNA cleavage assay
The cleavage assays were carried out with 1 pmole of 5' 32P-end labeled DNA substrate and 5–10 pmoles of topoisomerase I or Top67 in 10 μl of the buffer used for the gel mobility shift assay. After incubation at 37°C for up to 20 min, an equal volume of 90% formamide, 10 mM KOH, 0.25% bromophenol blue and 0.25% xylene cyanol was added to stop the reactions. The samples were analyzed by electrophoresis in a 12% sequencing gel followed by autoradiography.
Salt and Mg2+ induced reversal of cleavage
The conditions were modified from those described previously [14]. The cleavage reactions were incubated at 37°C for 5 min and then divided into three aliquots. The cleavage products were trapped in one aliquot by the addition of SDS to 1%. NaCl (1 M) alone or NaCl with MgCl2 (4 mM) were added to the other aliquots followed by further incubation at 37°C for up to 30 min before the addition of SDS. The products were analyzed as described for the cleavage reactions.
Catenation of nicked DNA circles
The catenation reaction was carried out with 1.4 μg of nicked PM2 phage DNA in 20 μl of 10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 10 mM KCl, 10 mM MgCl2. After incubation at 37°C for up to1 h, the reactions were stopped with the addition of 1% SDS and 50 mM EDTA. The products were analyzed as described for the relaxation assay.
References
Champoux JJ: DNA topoisomerases. Annu. Rev. Biochem. 2001, 70: 369-413. 10.1146/annurev.biochem.70.1.369.
Tse-Dinh YC: Bacterial and archeal type I topoisomerases. Biochim. Biophys. Acta. 1998, 1400: 19-27. 10.1016/S0167-4781(98)00125-0.
Drlica K: Control of bacterial DNA supercoiling. Mol. Microbiol. 1992, 6: 425-433.
Lima CD, Wang JC, Mondragon A: Three dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994, 367: 138-146. 10.1038/367138a0.
Tse-Dinh YC, Beran-Steed RK: Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J. Biol. Chem. 1988, 263: 15857-15859.
Ahumada A, Tse-Dinh YC: The Zn(II) binding motifs of Escherichia coli DNA topoisomerase I is part of a high-affinity DNA binding domain. Biochem. Biophys. Res. Comm. 1998, 251: 509-514. 10.1006/bbrc.1998.9500.
Grishin NV: C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J. Mol. Biol. 2000, 299: 1165-1177. 10.1006/jmbi.2000.3841.
Kirkegaard K, Pflugfelder G, Wang JC: The cleavage of DNA by type I DNA topoisomerases. Cold Spring Harbor Symp. Quant. Biol. 1984, 49: 411-419.
Tse YC, Kirkegaard K, Wang JC: Covalent bonds between protein and DNA: Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J. Biol. Chem. 1980, 255: 5560-5565.
Dean FB, Krasnow MA, Otter R, Matzuk MM, Splegler SJ, Cozzarelli NR: Escherichia coli type-1 topoisomerases: Identification, mechanism and role in recombination. Cold Spring Harbor Symp. Quant. Biol. 1982, 47: 769-777.
Lima CD, Wang JC, Mondragon A: Crystallization of a 67 kDa fragment of Escherichia coli DNA topoisomerase I. J. Mol. Biol. 1993, 232: 1213-1216. 10.1006/jmbi.1993.1474.
Tse-Dinh YC: Biochemistry of bacterial type I DNA topoisomerases. Adv. Pharmacology. 1994, 29A: 21-37.
Depew RE, Liu LF, Wang JC: Interaction between DNA and Escherichia coli protein ω: The formation of a complex between single-stranded DNA and ω protein. J. Biol. Chem. 1978, 253: 511-518.
Chen SJ, Wang JC: Identification of active site residues in Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1998, 273: 6050-6056. 10.1074/jbc.273.11.6050.
Tse YC, Wang JC: E. coli and M. luteus DNA topoisomerase I can catalyze catenation or decatenation of double-stranded DNA rings. Cell. 1980, 22: 269-276.
Krasnow MA, Cozzarelli NR: Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J. Biol. Chem. 1982, 257: 2687-2693.
DiGate RJ, Marians KJ: Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988, 263: 13366-13373.
Srivenugopal S, Lockshon D, Morris DR: Escherichia coli DNA topoisomerase III: purification and characterization of a new type I enzyme. Biochemistry. 1984, 23: 1899-1906.
Hiasa H, Marians KJ: Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J. Biol. Chem. 1994, 269: 32655-32659.
Zhang HL, DiGate RJ: The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J. Biol. Chem. 1994, 269: 9052-9059.
Zhang HL, Malpure S, Li Z, Hiasa H, DiGate RJ: The role of the carboxyl-terminal amino acid residues in Escherichia coli DNA topoisomerase III-mediated catalysis. J. Biol. Chem. 1996, 271: 9039-9045. 10.1074/jbc.271.15.9039.
Liu LF, Wang JC: Interaction between DNA and Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1979, 254: 11082-11088.
Valee BL, Coleman JE, Auld DS: Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains. Proc. Natl. Acad. Sci. 1991, 88: 999-1003.
Berg JM: Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J. Biol. Chem. 1990, 265: 6513-6516.
Qiu H, Kodadek T, Giedroc DP: Zinc-free and reduced T4 gene 32 protein binds single-stranded DNA weakly and fails to stimulate UvsX-catalyzed homologous pairing. J. Biol. Chem. 1994, 269: 2773-2781.
Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE: DNA ligase III Is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. J. Biol. Chem. 1999, 274: 21679-21687. 10.1074/jbc.274.31.21679.
Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP: Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC). J. Mol. Biol. 1998, 283: 419-434. 10.1006/jmbi.1998.2098.
Li Z, Mondragon A, Hiasa H, Marians KJ, DiGate RJ: Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol. Microbiol. 2000, 35: 888-895. 10.1046/j.1365-2958.2000.01763.x.
Mondragon A, DiGate RJ: The structure of Escherichia coli DNA topoisomerase III. Structure. 1999, 7: 1373-1383.
Keck JL, Berger JM: Enzymes that push DNA around. Nat Struct. Biol. 1999, 6: 900-902. 10.1038/13260.
Tse-Dinh YC: Zinc(II) coordination in Escherichia coli DNA topoisomerase I is required for cleavable complex formation with DNA. J. Biol. Chem. 1991, 266: 14317-14320.
Zhu CX, Qi H, Tse-Dinh YC: Mutation in Cys662 of Escherichia coli DNA topoisomerase I confers temperature sensitivity and change in DNA cleavage specificity. J. Mol. Biol. 1995, 250: 609-616. 10.1006/jmbi.1995.0402.
Viard T, Lamour V, Duguet M, de la Tours CB: Hyperthermophilic topoisomerase I from Thermotoga maritima. A very efficient enzyme that functions independently of zinc binding. J. Biol. Chem. 2001, 276: 46495-46503. 10.1074/jbc.M107714200.
Bhaduri T, Bagui TK, Sikder D, Nagaraja V: DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J. Biol. Chem. 1998, 273: 13925-13932. 10.1074/jbc.273.22.13925.
Suerbaum S, Brauer-Steppkes T, Labigne A, Cameron B, Drlica K: Topoisomerase I of Helicobacter pylori: juxtaposition with a flagellin gene (flaB) and functional requirement of a fourth zinc finger motif. Gene. 1998, 210: 151-161. 10.1016/S0378-1119(98)00065-1.
Beran-Steed RK, Tse-Dinh YC: The carboxyl terminal domain of E. coli DNA topoisomerase I confers higher affinity to DNA. Proteins Struct. Funct. Genet. 1989, 6: 249-258.
Caron PR: Compendium of DNA topoisomerase sequences. Methods Mol. Biol. 1999, 94: 279-316. 10.1385/1-59259-259-7:279.
Wilson TM, Chen AD, Hsieh T: Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J. Biol Chem. 2000, 275: 1533-1540. 10.1074/jbc.275.3.1533.
Masse E, Drolet M: Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 1999, 274: 16659-16664. 10.1074/jbc.274.23.16659.
Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID: The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol Chem. 2000, 275: 9636-9644. 10.1074/jbc.275.13.9636.
Zhu CX, Tse-Dinh YC: Overexpression and purification of bacterial DNA topoisomerase I. Methods Mol. Biol. 1999, 94: 145-151. 10.1385/1-59259-259-7:145.
Zumstein L, Wang JC: Probing the structural domains and function in vivo of Escherichia coli DNA topoisomerase I by mutagenesis. J. Mol. Biol. 1986, 191: 333-340.
Kivela HM, Mannisto RH, Kalkkinen N, Bamford DH: Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology. 1999, 262: 364-374. 10.1006/viro.1999.9838.
Wang JC: Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J. Mol. Biol. 1974, 87: 797-816.
Sternglanz R, DiNardo S, Voelkel KA, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang JC: Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc. Natl. Acad. Sci. USA. 1981, 78: 2747-51.
Acknowledgements
This work was supported by a grant (GM54226) to Y.T. and a predoctoral fellowship (GM17315) to A.A. from NIGMS, HHS. We thank Chang-Xi Zhu for preparation of topoisomerase I.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
Author 1 (A.A.) carried out all the experiments except the catenation assay. Author 2 (Y.T.) conceived of the study, participated in its design and coordination and carried out the catenation assay. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Ahumada, A., Tse-Dinh, YC. The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I. BMC Biochem 3, 13 (2002). https://doi.org/10.1186/1471-2091-3-13
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2091-3-13