Abstract
Background
The polymorphic species Candida albicans is the major cause of candidiasis in humans. The secreted aspartyl proteinases (Saps) of C. albicans, encoded by a family of 10 SAP genes, have been investigated as the virulent factors during candidiasis. However, the biological functions of most Sap proteins are still uncertain. In this study, we applied co-culture system of C. albicans and THP-1 human monocytes to explore the pathogenic roles and biological functions of Sap proteinases.
Results
After 1 hr of co-culture of C. albicans strains and THP-1 human monocytes at 37°C, more than 60% of the THP-1-engulfed wild type and Δsap5 Candida cells were developing long hyphae. However, about 50% of THP-1-engulfed Δsap6 Candida cells were generating short hyphae, and more dead Candida cells were found in Δsap6 strain that was ingested by THP-1 cells (about 15% in Δsap6 strain vs. 2 ~ 2.5% in SC5314 and Δsap5 strains). The immunofluorescence staining demonstrated that the Sap6 is the major hyphal tip located Sap protein under THP-1 phagocytosis. The sap6-deleted strains (Δsap6, Δsap4/6, and Δsap5/6) appeared slower growth on Congo red containing solid medium at 25°C, and the growth defect was exacerbated when cultured at 37°C in Congo red or SDS containing medium. In addition, more proteins were secreted from Δsap6 strain and the β-mercaptoethanol (β-ME) extractable surface proteins from Δsap6 mutant were more abundant than that of extracted from wild type strain, which included the plasma membrane protein (Pma1p), the ER-chaperone protein (Kar2p), the protein transport-related protein (Arf1p), the cytoskeleton protein (Act1), and the mitochondrial outer membrane protein (porin 1). Moreover, the cell surface accessibility was increased in sap6-deleted strains.
Conclusion
From these results, we speculated that the cell surface constitution of C. albicans Δsap6 strain was defect. This may cause the more accessible of β-ME to disulfide-bridged cell surface components and may weaken the resistance of Δsap6 strain encountering phagocytosis of THP-1 cells. Sap6 protein displays a significant function involving in maintenance the cell surface integrity.
Similar content being viewed by others
Background
Candida species are members of human normal microflora that reside in oral cavity, gastrointestinal tract, female genitalia and skin. Depending on the physiological status of the hosts, Candida species may convert from the commensally state to the pathogenic one and may cause disease from mucocutaneous superficial infection to systemic disseminated invasive candidiasis [1–3]. Among the members of genus Candida, the polymorphic species Candida albicans is the major contributor of candidiasis in humans [4, 5]. For surviving under various physiological stress of human host, C. albicans could transform between yeast, pseudohyphae and true hyphae in response to the environmental change [6].
For many microorganisms, including C. albicans, cell wall is the first line to get in touch with host cells and also provides defense to against attacks from the host immune system. Besides, the cell wall components would be modified during morphogenetic programs to cope with the changes in environmental conditions [7–9]. Therefore, the cell wall plays important roles in maintenance the integrity and homeostasis of microorganisms. Cell wall proteins of C. albicans are in general highly mannosylated (mannoproteins) and enriched in the outer surface. They are attached mostly to short chains of β-1,6-glucan, to chitin via β-1,6-glucan, or directly to chitin, and to β-1,3-glucan in a lesser extent. Three types of covalently bound cell wall proteins in C. albicans have been described, including: the proteins bound to β-1,6-glucans through a glycosylphosphatidylinositol (GPI) moiety, the Pir (proteins with internal repeats) proteins attached to β-1,3-glucan by unknown alkali-sensitive bonds (possibly O-glycosidic linkages), and the proteins retained by disulfide bridges which can be extracted by treatment with reducing agents such as β-mercaptoethanol (β-ME) or dithiothreitol [10–13].
The model yeast Saccharomyces cerevisiae has been used extensively for study the fungal cell wall biogenesis and cell wall integrity. Numerous molecules have been identified to participate in cell wall construction and cell wall integrity signaling [14–16], including a group of yapsin family proteins. In S. cerevisiae, the members of yapsin family are five glycosylphosphatidylinositol (GPI)-linked aspartyl proteinases [17]. In C. albicans, ten secreted aspartyl proteinase (Sap 1 ~ 10) were identified and categorized to a Sap protein family [18–22], which have been investigated as the virulent factors during candidiasis [2, 23, 24]. Among the ten Sap proteins, Sap9 and Sap10 contain the C-terminal GPI-linked sequences which make them to be the yapsin homologues of C. albicans[22, 25]. Studies have revealed that Sap9 and Sap10 are C. albicans cell surface-associated proteinases which cleave the covalently linked cell wall proteins [22, 26] and involved their functions in maintenance the cell wall integrity and mediation the interaction between C. albicans with human epithelial cells and neutrophils [22, 27].
In addition to SAP9 and SAP10, expression of SAP1 to SAP6 have been extensively approached and demonstrated that SAP1 ~ 3 were mainly expressed in yeast form C. albicans and SAP4 ~ 6 were hypha-associated expression [2]. Many studies revealed that Sap proteins of C. albicans are virulent factors during candidiasis [2, 23, 24] but some studies reflect that Sap proteins are not essential for pathogenesis of C. albicans[28–30]. However, the precise biological functions of most Sap proteinases in C. albicans are still uncertain. In this study, we co-cultured the C. albicnas and THP-1 human monocytes to examine the hyphae development and escape behavior of different sap-null mutants when suffered phagocytosis. We demonstrated that Sap6 involved in the maintenance the cell surface integrity of C. albicans.
Methods
Strains and media
The C. albicans strains used in this study were listed in Table 1[22, 26, 31–35]. Strains were grown on/in YPD (1% yeast extract, 2% peptone, 2% glucose) complex medium. YP (1% yeast extract, 2% peptone) medium containing 0.1% glucose was used for induction of hyphae development. All media were added 40 mg of uridine per liter to minimize the effect of URA3 gene [36]. C. albicans strains were cultured at 25°C to maintain the yeast form and incubated at 37°C for hyphae induction.
Polyclonal antibody preparation
The construction and preparation of recombinant Sap proteins for generation of polyclonal antibodies have been described [37]. Because the highly conserved protein sequences between C. albicans and S. cerevisiae, we took advantage of several antibodies that were generated by using recombinant proteins of S. cerevisiae as antigens to recognize the homologues in C. albicans. The detailed properties of antibodies used in this study were listed in Table 2.
C. albicans protein isolation and Western blot analysis
For total protein isolation, Candida cells were suspended in 200 μl of HEK solution (HEPES, pH7.4, 10 mM; EDTA 5 mM; KCl 50 mM) and added equal volume of glass beads. After a vigorous vortex for 10 min to break cells, 700 μl of HEK solution was added and incubated on ice for 10 min with occasional vortex. After centrifugation at 2,000 g for 5 min, the supernatant proteins were precipitated by 10% trichloroacetic acid (TCA). After centrifugation and washed with ddH2O, the protein pellet was suspended in 2× protein sample buffer and adjust pH value by 2 M Tris base, then incubated at 95°C for 10 min and stored at -20°C for further use. Western blotting was described previously [37].
Extraction of cell wall associated components
C. albicans strains were cultured overnight in YPD medium at 25°C. Cells were harvested and transferred into YPD or YP medium containing 0.1% glucose, with initial density of OD600 = 1/ml, and cultured at 25°C (yeast form) or 37°C (hyphal form) for 3 hr with shaking. Cells were spun down, washed once with ddH2O, then suspended in extraction buffer (Tris–HCl, pH 8.8, 20 mM; KCl 50 mM; β-mercaptoethanol 1%, v/v). After rocking at 37°C for 30 min, cells were harvested and the medium components were precipitated by 10% TCA. The precipitate was harvested by centrifugation at 20,000 g, 4°C, for 10 min. The pellet of precipitate was washed once by ddH2O then suspended in 2× protein sample buffer and adjust pH value by 2 M Tris base. After incubation at 95°C for 10 min, the components were subjected to Western blot analysis [12, 37].
Co-culture of C. albicans with THP-1 human monocytes
The THP-1 human monocytic cell line [38] is maintained in RPMI1640 with 10% fetal bovine serum (RPMI-FBS) at 37°C in a humidified chamber containing 5% CO2. For co-culture, THP-1 cells were cultured in the 10-cm dishes for 2 days, then suspended cells in fresh RPMI-FBS and incubated at 37°C for 10 min before co-cultured with Candida. About 2×106Candida cells were co-cultured with 2×105 THP-1 cells in 1.5 ml of RPMI-FBS in a 2 ml microcentrifuge tube at 37°C incubator for indicated times with gentle rocking [38].
Immunofluorescence staining of Sap proteins on hypha surface
C. albicans and THP-1 cells were co-cultured at 37°C for 30 min, then THP-1-engulfed Candida cells were harvested by low speed centrifugation and re-suspended in RPMI-FBS and incubated at 37°C for further 30 min. Co-cultured cells were harvested and suspended in PBS and loaded on poly-lysine coated cover glasses. The coated cells were fixed by 3.7% of formaldehyde in PBS for 15 min and were permeated by 0.2% TritonX100 for 3 min. After blocking, cells were incubated with anti-Sap6 antibody (1:800-dilution) for 90 min. The detailed procedure has been described [39].
RNA preparation and reverse transcription-polymerase chain reaction
Total RNA of Candida cells was isolated by hot acid phenol method [40]. Before reverse transcription, 2 μg of total RNA was treated by DNaseI (Invitrogen). The cDNA was generated by SuperScriptIII (Invitrogen) with oligo-(dT)12–18 as primer. The expression of SAP genes was further identified by PCR using specific primers [35].
Results
The characteristics of engulfed C. albicans in THP-1 human monocytes
The hypha-associated expression of SAP4-6 genes has been investigated as the potent virulent factors in mouse model of systemic candidiasis [34, 41]. However, the biological functions of these Sap proteinases are still uncertain. The environment of systemic infection is too complicate to dissect protein functions of pathogens. Because macrophages may be the first encountered host defense cells during the invasive process of pathogens, we applied co-culture of C. albicans and THP-1 human monocytes to evaluate the pathogenic roles of Sap proteinases. After co-culture of C. albicans and THP-1 cells at 37°C for 1 hr, we inspected the status of C. albicans that engulfed by THP-1 cells. The microscopy showed that one or more C. albicans could be ingested by one THP-1 cell, and C. albicans could be induced to develop filamentous growth within THP-1 cells. The cell shape of THP-1 cells appeared extended by the elongated hyphae of ingested Candida cells. Some elongated hyphae could eventually burst the THP-1 cells that seemed to kill the monocytes, but some Candida cells would likely to be killed by monocytes that appeared as hollow or dense images that mostly stayed in the yeast- or germ tube-form within the THP-1 cells (Figure 1).
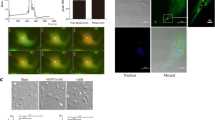
Sap6 participates in the breakout process of C. albicans from phagocytosis of THP-1 monocytes. Candida cells and THP-1 cells were co-cultured at 37°C for 1 hr. Then cells were harvested and prepared for microscope inspection. Thirty views of microscope for each sample were subjected to dissect and calculate the growth characters of co-cultured Candida cells. SC5314: the wild type strain; Δsap5: the SAP5 gene deleted mutant; Δsap6: the SAP6 gene deleted mutant; short hypha: filament </= two mother cell length; long hypha: filament > two mother cell length. *: the difference between Δsap6 strain and wild type/Δsap5 strains is considered to be statistically significant (t test, P value < 0.01).
Sap6 participates in the breakout process of C. albicans from phagocytosis
Because the Sap5 and Sap6 have been identified as the major expressed Sap proteins during hyphae development, we further characterized the properties of wild type strain SC5314, Δsap5 and Δsap6 mutants that were engulfed in THP-1 cells. We randomly took thirty views of each co-cultured samples and set several criteria to dissect the growth of C. albicans strains engulfed in THP-1 cells. The results revealed (Figure 1) that after 1 hr of co-cultur with THP-1 cells, most THP-1-engulfed SC5314 and Δsap5 cells were mostly developed long hyphae (about 60%). However, about 50% of THP-1-engulfed Δsap6 cells generated short hyphae, and more dead cells were found in THP-1-engulfed Δsap6 cells (about 15% in Δsap6 strain vs. 2 ~ 2.5% in SC5314 and Δsap5 strains). In addition, more protruding hyphae which pierced the THP-1 cells were found in THP-1-engulfed SC5314 and Δsap5 cells (about 12 ~ 15% in SC5314 and Δsap5 strains vs. 4% in Δsap6 strain). Hence, the Δsap6 mutant seemed with certain extent of defect in struggling of breakout from phagocytosis of THP-1 cells, although its hyphae formation is as efficient as other strains when cultured in the hypha-inducing media without THP-1 co-culture.
Sap6 is distributed on hyphal tips of THP-1 engulfed C. albicans
Since the THP-1-engulfed Δsap6 cells exhibited slower hypha-extension, we further identified the expression of Sap proteins in C. albicans co-cultured with THP-1 monocytes by immunofluorescence staining using polyclonal anti-Sap6 antibody. The results demonstrated (Figure 2) that the Sap proteins could be detected on the hyphal distal-end of THP-1 engulfed C. albicans. Although the Sap5 is the most abundant secreted aspartyl proteinase in hypha-form C. albicans, the fluorescent signal still appeared on the hyphal distal-end of THP-1 ingested Δsap5 cells. However, fluorescent signal was vanished from the hyphal surface of THP-1 engulfed Δsap6 strain, suggesting that under phagocytosis the Sap6 is the main hyphal-tip located Sap protein.
Sap6 is distributed on hypha-tip of THP-1 engulfed C. albicans . Co-cultured cells were harvested and subjected to immunofluorescence staining. Overlay: the fluorescence image was overlaid with the phase contrast image. SC5314: the wild type strain; Δsap2: the SAP2 gene deleted mutant; Δsap5: the SAP5 gene deleted mutant; Δsap6: the SAP6 gene deleted mutant.
The Δsap6 strain displayed growth defect in media with cell wall attack components
Meanwhile, we found when picking the C. albicans cells from colonies by pipette tip, the Δsap6 mutant appeared more liquefied and clammy than other strains; hence we speculated that the cell surface characteristics of Δsap6 mutant may be different from wild type and other strains. Because the outer layer of C. albicans cells is cell wall, we examined the growth of C. albicans strains on solid media of YPD and YPD containing Congo red to verify the cell wall integrity of Candida cells. All tested strains demonstrated similar growth rate on YPD medium at 25°C (Figure 3A). However, sap6-null strains (Δsap6, Δsap4/6, and Δsap5/6) appeared slower growth on Congo red containing medium at 25°C and displayed more severe growth defect when cultured at 37°C. The strain which contained one copy of re-integrated SAP6 gene seemed to present a little rescue of growth defect on Congo red plate that cultured at 25°C but there was no observable effect at 37°C (Figure 3A). The mRNA expression level of re-integrated SAP6 single-copy gene was approximately one-half of the wild type strain expression level in both yeast and hyphae form (Figure 3B), suggesting that the expression level of SAP6 is important to fulfill its biological function.
The expression level of SAP6 is related to its function. (A) The Δsap6 strains display growth defect in media with cell wall attack components. C. albicans strains were spotted on solid medium of YPD or YPD containing Congo red to verify the cell wall integrity of Candida cells. (B) Detection of SAP6 expression by reverse transcription-PCR analysis. The YPD cultured or co-cultured cells were harvested after 30 min incubation at 25°C or 37°C, respectively. Total RNA was isolated and subjected to reverse transcription and PCR analysis.
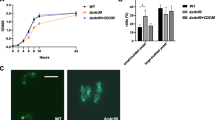
In addition, we cultured the C. albicans strains at 25°C in liquid rich medium or YPD rich medium containing 0.06% SDS (Figure 4A), the tested strains appeared almost the same growth rate in respective media. However, when the C. albicans strains were cultured at 37°C, the Δsap6 strain displayed evidently slower growth rate in medium containing 0.06% SDS (Figure 4B) and the single copy SAP6 gene re-integrated strain did not displayed growth rescue (data not shown). This result is consistent with the Congo red plate assay which revealed the growth defect was more sever when sap6-null strains were cultured in media with cell wall attack components at 37°C. From these results, we suggested that the cell wall constitution may be deficient in Δsap6 strain.
The Δ sap6 strain is more sensitive to SDS under hypha-induced condition. Candida strains were cultured in rich medium or rich medium contained 0.06% SDS at 25°C (A) or 37°C (B), respectively. Growth was measured at the specified time points. w/o: without. *: the difference between Δsap6 strain and wild type/Δsap5 strains is considered to be statistically significant (t test, P value < 0.05).
The accessibility of cell surface was increased in sap6-null strains
According to the cell wall constitution, wall components could be released or extracted by chemical agents. We applied the β-mercaptoethanol (β-ME) to extract surface proteins of C. albicans strains because this is a simple and efficient method to get a portion of outer layer proteins from C. albicans[10, 13].
The SDS-PAGE analysis revealed that in both yeast-form and hypha-form, the β-ME extractable proteins from Δsap6 mutant were evidently more than that extracted from wild type SC5314 strain (Figure 5A, upper panel). In addition, plenty of proteins also could be extracted by β-ME from a definite cell wall defect Δsap9 mutant [22], and the β-ME-extractable property of Δsap10 mutant was similar to wild type strain (Figure 5A, upper panel). The Western blotting demonstrated that plasma membrane protein (Pma1p), ER-chaperone protein (Kar2p), protein transport-related protein (Arf1p), cytoskeleton protein (actin, Act1), and trace of mitochondrial outer membrane protein (porin 1) could be detected in β-ME-extractable fraction, especially more abundant in the extracts of Δsap6 and Δsap9 strains (Figure 5A).
The surface accessibility is increased in sap6 -null strains. (A) More proteins can be extracted by β-ME from Δsap6 and Δsap9 strains. The plasma membrane protein (Pma1p), ER-chaperone protein (Kar2p), protein transport-related protein (Arf1p), cytoskeleton protein (actin, Act1), and mitochondrial outer membrane protein (porin 1) were detected by specific polyclonal antibodies. (B) Immunofluorescence staining was applied to detect the actin without treating Candida cells into spheroplast. Strains with sap6-deletion (Δsap6, Δsap4/6, and Δsap5/6) and Δsap9 mutant were more accessible to interact with Alexa Fluor™ 488 labeled anti-Act1 antibody. Anti-Mann: anti-mannan antibody.
Since the actin could be extracted from cells by β-ME treatment and some actin filaments should close to the inner surface of plasma membrane, we applied immunofluorescence staining to detect the actin without treating Candida cells into spheroplast. The result showed that sap6-null strains (Δsap6, Δsap4/6, and Δsap5/6) and Δsap9 strain were more accessible to interact with Alexa Fluor™ 488 labeled anti-Act1 antibody (Figure 5B), suggesting the surface accessibility was increased in sap6-null strains and Δsap9 strain.
Because the similar properties of Δsap6 and Δsap9 strains, we speculated that the cell surface constitution of Δsap6 strain was defect and gave rise to more accessible of β-ME to disulfide-bridged surface proteins; in addition, more proteins may be transported toward the cell surface to cope with the wall deficiency.
Discussion
In this study, we attempted to explore the biological function of secreted aspartyl proteinases in C. albicans. Fortunately, in the co-culture system of C. albicans and THP-1 human monocytes, we found the discrepancy of hyphae development between Δsap6 strain and other tested strains during phagocytosis. Substantially, the Δefg1 mutant, which exhibited severe deficiency of filamentous growth in many cultivated conditions, could almost not generate hyphae within THP-1 cells (data not shown); and a moderate hypha-deficient mutant (Δcph1) appeared delayed hyphae formation during phagocytosis (data not shown). In addition, the cell wall deficient Δsap9 strain [22] also exhibited poor hyphae development within THP-1 cells (data not shown). Therefore, co-culture of C. albicans and THP-1 human monocytes may apply as an efficient method for evaluation the potential factors relating to the invasiveness of C. albicans prior to examination by murine model.
A study using single and double null mutants of sap genes demonstrated that C. albicans strains, as long as with sap6-deletion, exhibited significantly reduced ability to invade and damage parenchymal organs in murine-model, although the hyphae development was normal and other Sap proteinases were still expressed [35]. Moreover, a study of murine keratitis also revealed that Sap6 is important for the pathogenesis of C. albicans keratitis [42]. In our co-culture system of C. albicans and THP-1 human monocytes, the protein level of secreted Sap5 was far more than the secreted Sap6 (Buu, unpublish), however, reduced capability of filamentous growth was found in Δsap6 strain of C. albicans within THP-1 monocytes. Besides, immunofluorescence staining revealed the Sap6 was the main Sap protein located on hyphal tips. These results highlight that the Sap6 may have distinct biological function involving in pathogenesis of C. albicans.
A class of GPI-anchored aspartyl proteinases known as fungal yapsins was firstly identified in Saccharomyces cerevisiae[25, 43, 44]. Yapsins in S. cerevisiae play important role in maintenance of cell wall integrity [17, 25]. C. albicans has ten secreted aspartyl proteinases but only Sap9 and Sap10 have been characterized as the homologues of yapsins for they possessed the GPI-modification signal in their C-terminal peptide sequences [22, 25]. Yps1, a plasma membrane reside yapsin of S. cerevisiae, has been investigated to be targeted to the vacuole and involving in the Golgi-associated proteolysis [45]. In addition, the IFF gene family of C. albicans encodes cell wall-related proteins. In this IFF gene family a secreted protein Iff11, which differs from other IFF family members in lacking a GPI anchor, has been investigated to play a role in cell wall organization, and presumed that the Iff11 may perform its function as it was transported through the protein secretory pathway [46]. The secreted Sap6 protein of C. albicans is also delivered through the conserved protein secretory pathway to the cell surface. Since the Sap6 is a proteinase, during transport through the secretory pathway it is possible that Sap6 provides cleavage-modification to maturate other co-delivered proteins which have main effect to establish the cell wall function.
In addition, S. cerevisiae Yps1 has been identified to cleave the extracellular inhibitory portion of the plasma membrane sensor protein Msb2p which is association with Sho1p for activation the Cdc42p-dependent MAPK pathway that controls filamentous growth and osmo-adaptive responses of S. cerevisiae[47]. In C. albicans, Msb2p is the sensor of defects in cell wall glycostructures and, associated with Sho1p, transmits the defective glycosylation signals to Cek1 MAP kinase pathway which functions in maintenance cell wall integrity [48]. Cleavage of Msb2p in C. albicans is also found to occur but is not performed by yapsin homologue Sap9 and Sap10, and Golgi-resident serine proteinase Kex2 is not involved as well [49]. A recent study revealed that Sap8 is a potent factor to be the major regulator of Msb2 processing in C. albicans[50]. These studies offer the hints that some physiological regulation processes are not exactly the same between S. cerevisiae and C. albicans, and there may have other unidentified cell surface-resident molecules also require further processing to exert their proper function when cells experience many different circumstances [51]. The Sap6 protein, though lacking a GPI-anchored moiety, may possibly retain on or associate with the cell surface constitution after it is secreted from the C. albicans and executes its proteinase activity on some surface molecules which may turn into active and participate in preservation the cell surface function.
Conclusion
This study demonstrates that although Sap4, Sap5, and Sap6 have high identity in their DNA and protein sequences, Sap6 displays a significant function involving in maintenance the cell surface integrity. Hence, secreted Sap6 is able to be a member of cell surface-modifying enzymes. The precise molecular mechanism of Sap6 will be further characterized that may help us to realize the various functions of Sap proteinases.
Abbreviations
- SAP:
-
Secreted aspartyl proteinase
- GPI:
-
Glycosylphosphatidylinositol
- β-ME:
-
β-mercaptoethanol
- TCA:
-
Trichloroacetic acid.
References
Chen Y-C, Chang S-C, Tai H-M, Hsueh P-R, Luh K-T: Molecular epidemiology of Candida colonizing critically ill patients in intensive care untis. J Formos Med Assoc. 2001, 100: 791-797.
Naglik J, Albrecht A, Bader O, Hube B: Candida albicans proteinases and host/pathogen interactions. Cell Microbiol. 2004, 6: 915-926. 10.1111/j.1462-5822.2004.00439.x.
Segal E: Candida, still number one- what do we know and where are we going from there?. Mycoses. 2005, 48: 3-11.
Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP: Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999, 29: 239-244. 10.1086/520192.
Ruan S-Y, Hsueh P-R: Invasive Candidiasis: an overview from Taiwan. J Formos Med Assoc. 2009, 108: 443-451. 10.1016/S0929-6646(09)60091-7.
Whiteway M, Bachewich C: Morphogenesis in Candida albicas. Annu Rev Microbiol. 2007, 61: 529-553. 10.1146/annurev.micro.61.080706.093341.
Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP: Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2006, 2: 0204-0210.
Biswas S, Van Dijck P, Datta A: Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007, 71: 348-376. 10.1128/MMBR.00009-06.
Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP: An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010, 6: e1000752-10.1371/journal.ppat.1000752.
Pitarch A, Sánchez M, Nombela C, Gil C: Sequential fractionation and two- dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol Cell Proteomics. 2002, 1: 967-982. 10.1074/mcp.M200062-MCP200.
de Groot PW, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, de Koster C, Klis FM: Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot Cell. 2004, 3: 955-965. 10.1128/EC.3.4.955-965.2004.
Ruiz-Herrera J, Elorza MV, Valentín E, Sentandreu R: Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006, 6: 14-29. 10.1111/j.1567-1364.2005.00017.x.
Thomas DP, Pitarch A, Monteoliva L, Gil C, Lopez-Ribot JL: Proteomics to study Candida albicans biology and pathogenicity. Infect Disord Drug Targets. 2006, 6: 335-341. 10.2174/187152606779025815.
Levin DE: Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005, 69: 262-291. 10.1128/MMBR.69.2.262-291.2005.
Klis FM, Boorsma A, De Groot PWJ: Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006, 23: 185-202. 10.1002/yea.1349.
Lesage G, Bussey H: Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006, 70: 317-343. 10.1128/MMBR.00038-05.
Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS: Yapsins are a family of aspartyl Proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell. 2005, 4: 1364-1374. 10.1128/EC.4.8.1364-1374.2005.
White TC, Miyasaki SH, Agabian N: Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993, 175: 6126-6133.
Hube B, Monod M, Schofield DA, Brown AJ, Gow NAR: Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994, 14: 87-99. 10.1111/j.1365-2958.1994.tb01269.x.
Monod M, Togni G, Hube B, Sanglard D: Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 1994, 13: 357-368. 10.1111/j.1365-2958.1994.tb00429.x.
Monod M, Hube B, Hess D, Sanglard D: Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology. 1998, 144: 2731-2737. 10.1099/00221287-144-10-2731.
Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, MacCallum D, Odds FC, Schafer W, Klis F, Monod M, Hube B: Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem. 2006, 281: 688-694.
Calderone RA, Fonzi WA: Virulence factors of Candida albicans. Trends Microbiol. 2001, 9: 327-335. 10.1016/S0966-842X(01)02094-7.
Naglik JR, Challacombe SJ, Hube B: Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003, 67: 400-428. 10.1128/MMBR.67.3.400-428.2003.
Gagnon-Arsenault I, Tremblay J, Bourbonnais Y: Fungal yapsins and cell wall: a unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res. 2006, 6: 966-978. 10.1111/j.1567-1364.2006.00129.x.
Schild L, Heyken A, de Groot PWJ, Hiller E, Mock M, de Koster C, Horn U, Rupp S, Hube B: Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans sap9 and Sap10. Eukaryot Cell. 2011, 10: 98-109. 10.1128/EC.00210-10.
Hornbach A, Heyken A, Schild L, Hube B, Loffler J, Kurzai O: The Glycosylphosphatidylinositol-Anchored Protease Sap9 modulates the Interaction of Candida albicans with human neutrophils. Infect Immun. 2009, 77: 5216-5224. 10.1128/IAI.00723-09.
Kretschmar M, Felk A, Staib P, Schaller M, Heb D, Callapina M, Morschhauser J, Schafer W, Korting HC, Hof H, Hube B, Nichterlein T: Individual acid aspartic proteinases (Saps) 1–6 of Candida albicans are not essential for invasion and colonization of the gastrointestinal tract in mice. Microb Pathog. 2002, 32: 61-70. 10.1006/mpat.2001.0478.
Lermann U, Morschhauser J: Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology. 2008, 154: 3281-3295. 10.1099/mic.0.2008/022525-0.
Correia A, Lermann U, Teixeira L, Cerca F, Botelho S, Gil da Costa RM, Sampaio P, Gartner F, Morschhauser J, Vilanova M, Pais C: Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect Immun. 2010, 78: 4839-4849. 10.1128/IAI.00248-10.
Gillum AM, Tsay EY, Kirsch DR: Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mu tations. Mol Gen Genet. 1984, 198: 179-182. 10.1007/BF00328721.
Fonzi WA, Irwin MY: Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993, 134: 717-728.
Hube B, Sanglard D, Odds FC, Hess D, Monod M, Schafer W, Brown AJ, Gow NAR: Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, an d SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997, 65: 3529-3538.
Sanglard D, Hube B, Monod M, Odds FC, Gow NAR: A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997, 65: 3539-3546.
Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schafer W, Hube B: Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun. 2002, 70: 3689-3700. 10.1128/IAI.70.7.3689-3700.2002.
Brand A, MacCallum DM, Brown AJP, Gow NAR, Odds FC: Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004, 3: 900-909. 10.1128/EC.3.4.900-909.2004.
Chen Y-C, Wu C-C, Chung W-L, Lee F-J S: Differential secretion of Sap4-6 proteins in Candida albicans during hyphae formation. Microbiology. 2002, 148: 3743-3754.
Barker KS, Liu T, Rogers PD: Coculture of THP-1 human mononuclear cells with Candida albicans results in pronounced changes in host gene expression. J Infect Dis. 2005, 192: 901-912. 10.1086/432487.
Buu L-M, Chen Y-C, Lee F-J S: Functional characterization of acetyl-CoA hydrolase, Ach1p, in Saccharomyces cerevisiae. J Biol Chem. 2003, 278: 17203-17209. 10.1074/jbc.M213268200.
Collart MA, Oliviero S: Supplement Unit 13.12. Preparation of yeast RNA. Current Protocols in Molecular Biology. Edited by: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1993, New York: John Wiley & Sons, Inc, 13.12.1-5
Kretschmar M, Hube B, Bertsch T, Sanglard D, Merker R, Schroder M, Hof H, Nichterlein T: Germ tubes and proteinase activity contribute to virulence of Candida albicans in murine peritonitis. Infect Immun. 1999, 67: 6637-6642.
Jackson BE, Wilhelmus KR, Hube B: The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci. 2007, 48: 3559-3565. 10.1167/iovs.07-0114.
Bourbonnais Y, Ash J, Daigle M, Thomas DY: Isolation and characterization of S. cerevisiae mutants defective in somatostatin expression: cloning and functional role of a yeast gene encoding an aspartyl protease in precursor processing at monobasic cleavage sites. EMBO J. 1993, 12: 285-294.
Ash J, Dominguez M, Bergeron JJM, Thomas DY, Bourbonnais Y: The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. J Biol Chem. 1995, 270: 20847-20854. 10.1074/jbc.270.35.20847.
Sievi E, Suntio T, Makarow M: Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic. 2001, 2: 896-907. 10.1034/j.1600-0854.2001.21205.x.
Bates S, de la Rosa JM, MacCallum DM, Brown AJP, Gow NAR, Odds FC: Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect Immun. 2007, 75: 2922-2928. 10.1128/IAI.00102-07.
Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ: Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 2008, 181: 1073-1081. 10.1083/jcb.200704079.
Ernst JF, Pla J: Signaling the glycoshield: maintenance of the Candida albicans cell wall. Int J Med Microbiol. 2011, 301: 378-383. 10.1016/j.ijmm.2011.04.003.
Szafranski-Schneider E, Swidergall M, Cottier F, Tielker D, Roman E, Pla J, Ernst JF: Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 2012, 8: e1002501-10.1371/journal.ppat.1002501.
Puri S, Kumar R, Chadha S, Tati S, Conti HR, Hube B, Cullen PJ, Edgerton M: Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One. 2012, 7: e46020-10.1371/journal.pone.0046020.
Cullen PJ: Post-translational regulation of signaling mucins. Curr Opin Struct Biol. 2011, 21: 590-596. 10.1016/j.sbi.2011.08.007.
Acknowledgments
The authors are grateful to Drs. Bernhard Hube, Dominique Sanglard, Gerald R. Fink, and William A. Fonzi for providing the strains of C. albicans, and thankful to Dr. Fang-Jen S. Lee for providing antibodies. We thank the facilities support at the 6th Core Laboratory, National Taiwan University Hospital. This work was supported by the National Science Council, Taiwan (NSC 98-2320-B-017-002).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LMB designed and manipulated the experiments and wrote the manuscript. YCC discussed the experimental design and results with LMB and participated in manuscript writing. Both authors read and approve the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Buu, LM., Chen, YC. Sap6, a secreted aspartyl proteinase, participates in maintenance the cell surface integrity of Candida albicans. J Biomed Sci 20, 101 (2013). https://doi.org/10.1186/1423-0127-20-101
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1423-0127-20-101