Abstract
Background
Schizophrenia is a complex disorder with involvement of multiple genes.
Methods
In this study, genome-wide screening for DNA copy-number variations (CNVs) was conducted for ten pairs, a total of 20 cases, of affected siblings using oligonucleotide array-based CGH.
Results
We found negative symptoms were significantly more severe (p < 0.05) in the subgroup that harbored more genetic imbalance (n ≧ 13, n = number of CNV-disrupted genes) as compared with the subgroup with fewer CNVs (n ≦ 6), indicating that the degree of genetic imbalance may influence the severity of the negative symptoms of schizophrenia. Four central nervous system (CNS) related genes including CCAAT/enhancer binding protein, delta (CEBPD, 8q11.21), retinoid × receptor, alpha (RXRA, 9q34.2), LIM homeobox protein 5 (LHX5, 12q24.13) and serine/threonine kinase 11 (STK11, 19p13.3) are recurrently (incidence ≧ 16.7%) disrupted by CNVs. Two genes, PVR (poliovirus receptor) and BU678720, are concordantly deleted in one and two, respectively, pairs of co-affected siblings. However, we did not find a significant association of this BU678720 deletion and schizophrenia in a large case-control sample.
Conclusions
We conclude that the high genetic loading of CNVs may be the underlying cause of negative symptoms of schizophrenia, and the CNS-related genes revealed by this study warrant further investigation.
Similar content being viewed by others
Background
Schizophrenia is a devastating mental disorder, and its etiology has yet to be fully elucidated. Genetic epidemiological studies have shown that schizophrenia is predominantly genetically determined and has a high heritability, with a multi-locus inheritance model [1]. Chromosomal abnormalities occurring in patients with schizophrenia may provide useful information for locating and fine mapping the relevant gene loci. This has been demonstrated by the identification of the potential vulnerability genes of proline dehydrogenase (PRODH) [2] and Disrupted-in-Schizophrenia 1 (DISC1) [3, 4] based on 22q11 micro-deletion syndrome and balance translocation (1;11) (q42.1;q14.3), respectively.
Although linkage studies in schizophrenia have provided some evidence of susceptible loci over many broad chromosomal regions, pinpointing causative gene mutations by conventional linkage strategy alone is problematic [5]. On the other hand, the resolution, typically ranging from 5 to 10 Mb [6, 7], of traditional cytogenetic techniques such as fluorescence in situ hybridization (FISH), comparative genomic hybridization (CGH), and spectral karyotyping (SKY)-FISH is limited, and submicroscopic aberrations (fewer than a few tens of kilobases) of genomic DNA may be impossible to identify using these methods. Due to the technical constraints, we believe that the importance of chromosomal anomalies that could be the major cause of schizophrenia may have been overlooked in the past.
One of the chromosomal alterations involving amplifications and deletions of genetic materials is referred to as a copy number variant (CNV). Because of the rapid development of molecular genetic tools, recent studies have demonstrated the presence of several CNVs within the human genome. Some genomic CNVs called copy number polymorphisms (CNPs) may not be pathogenic and simply contribute to human genetic diversity and individual variability in response to environmental stimuli [8–10]. On the other hand, some CNVs have proven to be associated with several human diseases, including cancer [11], intellectual disability [12–14], and autism [15]. These discoveries have encouraged investigators to study CNVs in complex disorders like schizophrenia. Using the array-based comparative genomic hybridization (array CGH) technology, many genome-wide surveys of CNVs implicated in schizophrenia have been completed [16–24]. Three large-scale studies [17, 19, 22] suggested that rare copy number alterations collectively are significant risk factors for this disease. These works also revealed that specific CNVs on chromosomes 1 and 15 were responsible for vulnerability to schizophrenia. In addition, Kirov's work [24] suggested that a deletion at 2p16.3 disrupting NRXN1 and a duplication at 15q13.1 spanning APBA2 may be implicated in schizophrenia. Even so, a large fraction of the overall genetic risk for schizophrenia remains unexplained.
The present study explored the CNVs in genomic DNA of familial schizophrenia, assuming that the higher genetic loading in schizophrenic families may reveal significant copy number aberrations. Thus, we studied co-affected siblings to highlight the influence of predisposing genetic components and expected to find concordant CNVs in them.
Methods
Recruitment of healthy controls
Healthy controls were recruited from the employees of the National Taiwan University Hospital. After signing informed consent, the individuals underwent a screening interview followed by blood withdrawal. Exclusion criteria for the healthy controls were: under age 30; diagnosed with psychiatric disorder, especially schizophrenia; having a history of diabetes mellitus (DM), major systemic disorder, or neurological disorder (e.g., epilepsy); mental retardation; facial dysmorphism; and clinical evidence of brain, trunk or limb anomalies.
Establishment of control genomic DNA pool
The control DNA pools were constructed by pooling equal amounts of DNA extracted from ten healthy men and ten healthy women. These normal genomic DNA pools were used as reference samples for array CGH analysis and real-time quantitative PCR.
Recruitment of schizophrenics
Patients for array CGH analysis were enrolled from the outpatient clinics of the Department of Psychiatry, National Taiwan University Hospital. The inclusion criteria were: a diagnosis of schizophrenia according to the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) [25] and confirmed by the Diagnostic Interview for Genetic Study (DIGS) and at least two siblings affected by schizophrenia in a given family. Patients affected with mental retardation, facial dysmorphysm, or clinical evidence of brain, trunk or limb anomalies were excluded. A total of ten pairs of schizophrenic siblings, a total of 20 cases, from ten unrelated Taiwanese families (A-J), were recruited. The mean age of the subjects was 30.6 years.
After the homozygous deletion of BU678720 was found in the initial 20 subjects, a different sample was recruited between 2003 and 2005 in Taiwan for genetic study. A total of 107 controls, 163 simplex schizophrenic patients (those from families with only one member affected with schizophrenia), and 72 multiplex schizophrenic patients (those from families having at least two affected siblings) were included in this study. All cases fulfilled the DSM-IV criteria for schizophrenia.
Assessment of clinical psychopathology for schizophrenia
Clinical symptoms were rated using the scale for the assessment of negative symptoms (SANS) [26] and the scale for the assessment of positive symptoms (SAPS) [27], both of which have demonstrated satisfactory reliability. According to SANS, the negative symptom score was the sum of scores for Affective Blunting, Alogia, Avolition-Apathy, and Anhedonia-Asociality. The Continuous Performance Test (CPT) [28] and Wisconsin Card Sorting Test (WCST) [29] were used for neuropsychological assessment of sustained attention and executive function, respectively. The assessment methods were described in our previous reports [28, 29].
Genomic DNA extraction
Genomic DNA was isolated from peripheral blood lymphocytes with the PureGene DNA Purification Kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer's instructions.
Array-based CGH (array CGH) experiment
A commercial oligonucleotide array (Human Genome CGH microarray 44B, Agilent Technologies, Palo Alto, CA, USA) was used for array-CGH analysis. Genomic DNA fragmentation, labelling and array hybridization were performed as previously described [30, 31]. Each array hybridization experiment was performed with differentially labelled gender-matched samples, one from the affected individual, and the other from the DNA control pool. To rule out probable CNPs in our ethnic group, two array hybridization experiments were performed using the male or female pooled control and commercial, normal, same-gender Caucasian samples (Promega, Madison, WI, USA).
Selection of high-confidence copy number alterations
Filtering procedures were applied to select qualified data sets for analysis. In total, 18 qualified arrays, exclusive of the samples for patients F2 and J2, were selected for further analysis. Aberrations were only considered if the aberration scores, automatically generated by Agilent CGH analysis software, were higher than 1.00 or lower than -1.00. The CNVs which exist within the control genome and are unlikely to be pathogenic were filtered out by comparison to the CNVs identified from the two reference arrays which were performed with normal male and female DNA pools in our laboratory. Additionally, the published CNVs listed in the Database of Genomic Variants [32] with relative high incidence in control subject were also excluded.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) [33] and are accessible through GEO series accession number GSE16930.
Examination of array CGH findings
In order to examine the copy number changes of those genes found in our array CGH analysis, we used the quantitative real-time PCR method. The detailed qPCR conditions were described by Lee [31]. The specific oligonucleotide primer pairs were selected from the Universal Probe Library (Roche Molecular Systems, Inc., Branchburg, NJ, USA). The ATPase, Ca++ transporting, plasma membrane 4 (ATP2B4, NM_001001396) was chosen as the reference. The fold change in gene copy number for a target gene is calculated by using the comparative ΔCT method as described previously [34]. In each experiment the samples were analyzed in triplicate. The primer information is provided as Supplementary Table 1 (Additional file 1).
In order to investigate the prevalence of homozygous deletion of the specific UB678720 allele revealed in this array CGH study in a different sample, we used PCR followed by electrophoresis. PCR was carried out with the same primers as those used for qPCR. Details of PCR conditions were as described elsewhere [31]. Amplified products were analyzed by on-chip electrophoresis using Agilent 2100 bioanalyzer and Agilent DNA 1000 LabChip kit (Agilent Technologies). Homozygous deletion of BU678720 was readily distinguishable by the presence of the 85 bp amplified fragment.
Data analysis methods
For array CGH analysis, the hybridized arrays were scanned and analyzed as previously described [30, 31]. Briefly, after washing, the hybridized arrays were immediately scanned at a resolution of 5 μ using an Agilent G2565BA DNA microarray scanner. The microarray images were analyzed using Agilent Feature Extraction software, version 8.1.1. Another custom analytical software package, Agilent CGH Analytics, version 3.4, was used for the subsequent data analysis. The locations of the copy number aberrations were calculated using the Aberration Detection Method 2 (ADM2) statistical algorithm. The ADM2 threshold was set at 9.0 to make an amplification or deletion determination. According to these settings, the aberration score was generated automatically for each altered locus.
The comparison of the negative symptoms between subgroups was calculated by using the Mann-Whitney U test. The comparison of the incidence of BU678720 homozygous deletion between multiplex and simplex families was analyzed by using the Genmod procedure with software SAS 9.1.
Results
Classification of patients with schizophrenia
A total of 379 loci disrupted by CNVs were found. These included 343 losses, 10 gains, and 26 with both losses and gains. Summaries for each patient are presented in Table 1. There were great variations in the number of loci affected by CNVs (range 0-318). Thus we classified patients into subgroups based on the number of loci with CNVs. There were six or fewer scattered loci disrupted by CNVs in subgroup I (A1, B2, C1, E1, F1, G1, G2, and J1), while subgroup II had up to 318 loci with CNVs (A2, B1, C2, D1, D2, E2, H1, H2, I1 and I2). We also noted that the patterns of CNVs showing deletions were more common than those showing amplifications. In addition, we observed that four sib-pair subjects (50%) were in different subgroups, while there were four sib-pair subjects (50%) in the same subgroup (one in subgroup I, three in subgroup II) (Table 1).
Comparison of psychopathological parameters
We compared the clinical features between subgroup I and subgroup II patients and examined whether the array CGH profiles were correlated with the phenotypes defined by psychopathological parameters of clinical symptoms and neuropsychological performance. We found that the negative symptom score was significantly higher (Mann-Whitney U = 14, n1 = 8, n2 = 9, p = 0.033, two-tailed) for subgroup II when compared to subgroup I (Table 1). However, there was no significant difference in the scores for delusions/hallucinations and disorganized symptoms dimensions. There were also no differences in the performance of sustained attention and executive function between these two subgroups.
Potential candidate genes revealed by array CGH
The genes related to CNS growth and development were referred to as CNS-related from a computer search of the relevant literature on PubMed [35]. For subgroup I patients, the three CNS-related aberrant loci including BTB (POZ) domain containing 8 (BTBD8, NM_183242, 1p22.1), paired-like homeobox 2b (PHOX2B, NM_003924, 4p12) and apoptosis-associated tyrosine kinase (AATK, NM_001080395, 17q25.3) were detected in patients A1, G2 and F1, respectively. It was noted that the copy number gain of AATK and copy number loss of PHOX2B were the only CNVs detected in patients F1 and G2, respectively.
Of the genes disrupted by CNVs identified in the subgroup II patients (10 subjects), a total of 16 genes with an incidence of at least 30% (3/10) were selected. The distributions of these highly recurrent CNVs are presented in Table 2. Four of these 16 target loci, genes encoding CCAAT/enhancer binding protein, delta (CEBPD, NM_005195), retinoid × receptor, alpha (RXRA, NM_002957), LIM homeobox protein 5 (LHX5, NM_022363), serine/threonine kinase 11 (STK11, NM_000455) are CNS-related. There are only two genes, PVR and BU678720, with familial incidence. Of these two genes, the BU678720 (BU678720) (function not yet known) had a concordant loss in both sib-pairs of families B and D (Figure 1) and accounted for the highest familial incidence (Additional file 2: Supplementary Table 2). In comparison with healthy controls determined by screening against the Database of Genomic Variants, the alteration incidences are rather high in schizophrenics for all 16 identified CNVs (Additional file 2: Supplementary Table 2, Table 2, Fig 1).
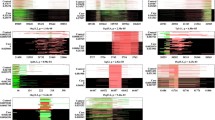
Concordant loss of BU678720 in sib-pairs of schizophrenics. (a) Array CGH profiles are shown for patients B1, B2, D1 and D2. The X axis marks the chromosome coordinate in megabases. The Y axis marks the hybridization ratio plotted in log2 scale. In these four patients, there is strong indication of a loss of BU678720 (Arrows). Graphics are produced by using Agilent software CGH Analytics version 3.4. (b) LOH analysis of BU678720. Gel view of the PCR products of BU678720 and GAPDH were shown for all affected sib-pairs. The specific amplified fragments of BU678720 and GAPDH were indicated. Graphics are generated by Agilent Bioanalyzer.
Examination of array CGH findings
Quantitative PCR analysis was performed for the four recurrent CNS-related genes, CEBPD, RXRA, LHX5, STK11, and the gene of BU678720 with a high incidence (two out of 8 pairs: 25%) of concordant loss in both sib-pairs. We analyzed the siblings for whom CNS-related CNVs were detected in at least one of the sib pairs. Comparisons between results from arrays and those from qPCR are summarised in Supplementary Figure 1 (Additional file 3). Deletions of BU678720 could be confirmed by qPCR, while, for others, the qPCR patterns were not perfectly consistent for array CGH results. For BU678720, the low qPCR signals suggested that the gene copy may be completely lost in sib pairs from families B and D.
Since homozygous deletion may have a more profound influence on regulation of gene expression than single copy deletion, we performed the PCR experiments on the ten pairs of affected siblings to detect a possible homozygous deletion in the four CNS-related candidate genes (CEBPD, RXRA, LHX5 and STK11) and the gene BU678720. Homozygous deletion was detected in the gene BU678720 in six individuals A1, B1, B2, D1, D2, and J1 (Fig. 1b), while no homozygous deletion was detected in the four CNS-related candidate genes. To assess whether the homozygous deletion cosegregated with schizophrenia in the four families, we recruited the first-degree relatives of families A, B, D and J to examine for homozygous deletion of BU678720. The results are depicted in Figure 2. Homozygous loss of BU678720 cosegregated with schizophrenia in families B and D. However, the phenomenon of cosegregation was not observed in families A and J. To further clarify the association between BU678720 and schizophrenia, we performed the PCR assay in 72 and 163 affected individuals from multiplex and simplex families, respectively, as well as in 107 controls. The homozygous deletion of BU678720 was detected in 7.4% of patients from simplex families, 11.1% of patients from multiplex families, and 8.4% of controls. Though the prevalence in the patients from the multiplex families was higher than that in controls and patients for simplex families, the comparisons did not attain a level of statistical significance (p = 0.098, df = 1).
Discussion
Schizophrenia is known as an etiologically diverse psychiatric disorder which exhibits both familial (hereditary) and nonfamilial (sporadic) patterns. To increase the possibility of discovering genomic numerical alterations that contribute to the genetic component of schizophrenia, we recruited 10 affected sibling pairs, a total of 20 affected subjects, for a genome-wide study with the aid of commercial CGH oligonucleotide array. There are conspicuous differences which discriminate our experimental design from those of similar recent investigations: predisposing genetic aberrations as the major causative factor have been emphasized in our study sample of ten affected sibling pairs, instead of sporadic patients without a positive family history of schizophrenia; and, the use of oligoarray (average spatial resolution approximately 35 Kb) instead of BAC array for array CGH has the potential to directly indicate the genes associated with schizophrenia.
The prevalence of concordant CNVs in pairs of co-affected siblings seems low. We found that only two genes, PVR and BU678720, had concordant deletions in both affected siblings with 12.5% (1/8) and 25% (2/8), respectively, of familial incidence. Using three different experiment designs of array CGH, qPCR and PCR, we found that the homozygous deletion of BU678720 cosegregated with schizophrenia in the two families. However, we did not find a statistically significant association between the homozygous deletion of BU678720 and schizophrenia in a larger case-control sample. We did find that the prevalence of homozygous deletion of this gene was higher in the multiplex patients, but at borderline significance. We cannot totally exclude the possibility of homozygous deletion of BU678720 in the pathogenesis of familial schizophrenia. The gains and losses of genomic material assessed by array CGH seemed not to run in multiplex families. The genetic etiology in multiplex family may be better explained by other factors.
By classifying patients into 2 subgroups according to the quantity of CNV-disrupted genes, we found a correlation between the clinical psychopathological manifestations and the total burden of CNVs in DNA content. The patients with more CNVs exhibited significantly more severe negative symptoms than did those with fewer CNVs. This observation may imply that nonspecific lesions in copy numbers of the somatic genome are a discriminative property among families with schizophrenia and have applicability in predicting an elevated risk for negative symptoms. It is also interesting to note that losses are more common than gains in the patterns of numerical aberrations detected. Several genomic aberration studies of neurological disorders agreed with the finding that more frequent copy number deletions might cause disease. However, the sample size for array CGH analysis is rather small in this pilot study, and the correlation between the number of CNVs and negative symptoms may be weakened by extending the number of cases. Thus, these preliminarily results should be treated with caution.
We did find four CNS-related candidate vulnerability genes in subgroup II patients with a higher number (≥ 13) of CNVs. It is worth noting that STK11, found to be copy-number lost in 30% of this subgroup, has been identified as a large-scale deletion in a patient with schizophrenia and Peutz-Jeghers syndrome [36]. RXRA showed an exceedingly high deletion rate in five of our patient cohorts (C2, D1, E2, H1, and I) by both array CGH and qPCR. Wallen-Mackenzie et al [37] have reported that RXRA may contribute to schizophrenia via interaction with Nurr1. Additionally, several relevant reports in the literature also support its involvement in schizophrenia [38–43]. The chromosomal region containing RXRA, 9q34, has been associated with schizophrenia [16]. This uniformity cross-validates our results and confirms the significance of RXRA deletion. Two other CNS-related candidate genes, CEBPD and LHX5 may be associated with schizophrenia, but the results of array CGH should be treated carefully in the absence of validation by other technologies.
We intended to examine the results of array CGH by using qPCR methodology, but we do not have positive results in this regard; however, the inefficiency in validating array CGH data by using qPCR methodology has also been reported previously [18]. The chromosomal distances between the array probes and primers for qPCR may account for the discrepancies between the results of array CGH and qPCR. This may also be due to the complexity of genome sequences that influence the specificity of primers of qPCR. Thus, we still can not exclude these four CNS-related genes found in this array CGH study as schizophrenia vulnerability genes.
Conclusions
In summary, our work further demonstrated that oligonucleotide array CGH is a useful platform for investigating the genomic aberrations of psychiatric disorders. We found the sum of altered gene dosage is coincident with severity of the negative symptoms of schizophrenia. Additionally, the CNS-related genes including CEBPD, RXRA, LHX5, and STK11 revealed by this pilot study may also provide entry points for further investigation.
Abbreviations
- (array CGH):
-
Array-based comparative genomic hybridization
- (CNVs):
-
copy number variations
- (CNPs):
-
copy-number polymorphisms.
References
Kringlen E: Twin studies in schizophrenia with special emphasis on concordance figures. Am J Med Genet. 2000, 97: 4-11. 10.1002/(SICI)1096-8628(200021)97:1<4::AID-AJMG2>3.0.CO;2-J.
Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, Lenane M, Robertson B, Wijsman EM, Rapoport JL, Gogos JA, Karayiorgou M: Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002, 99: 3717-3722. 10.1073/pnas.042700699.
Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ: Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000, 9: 1415-1423. 10.1093/hmg/9.9.1415.
Millar JK, Christie S, Anderson S, Lawson D, Hsiao-Wei Loh D, Devon RS, Arveiler B, Muir WJ, Blackwood DH, Porteous DJ: Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry. 2001, 6: 173-178. 10.1038/sj.mp.4000784.
Kato C, Petronis A, Okazaki Y, Tochigi M, Umekage T, Sasaki T: Molecular genetic studies of schizophrenia: challenges and insights. Neurosci Res. 2002, 43: 295-304. 10.1016/S0168-0102(02)00064-0.
Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992, 258: 818-821. 10.1126/science.1359641.
Macville M, Schrock E, Padilla-Nash H, Keck C, Ghadimi BM, Zimonjic D, Popescu N, Ried T: Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999, 59: 141-150.
Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C: Detection of large-scale variation in the human genome. Nat Genet. 2004, 36: 949-951. 10.1038/ng1416.
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME: Global variation in copy number in the human genome. Nature. 2006, 444: 444-454. 10.1038/nature05329.
Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M: Large-scale copy number polymorphism in the human genome. Science. 2004, 305: 525-528. 10.1126/science.1098918.
Zafarana G, Grygalewicz B, Gillis AJ, Vissers LE, Vliet van de W, van Gurp RJ, Stoop H, Debiec-Rychter M, Oosterhuis JW, van Kessel AG, Schoenmakers EF, Looijenga LH, Veltman JA: 12p-amplicon structure analysis in testicular germ cell tumors of adolescents and adults by array CGH. Oncogene. 2003, 22: 7695-7701. 10.1038/sj.onc.1207011.
Lugtenberg D, de Brouwer AP, Kleefstra T, Oudakker AR, Frints SG, Schrander-Stumpel CT, Fryns JP, Jensen LR, Chelly J, Moraine C, Turner G, Veltman JA, Hamel BC, de Vries BB, van Bokhoven H, Yntema HG: Chromosomal copy number changes in patients with non-syndromic × linked mental retardation detected by array CGH. J Med Genet. 2006, 43: 362-370. 10.1136/jmg.2005.036178.
Van Esch H, Hollanders K, Badisco L, Melotte C, Van Hummelen P, Vermeesch JR, Devriendt K, Fryns JP, Marynen P, Froyen G: Deletion of VCX-A due to NAHR plays a major role in the occurrence of mental retardation in patients with X-linked ichthyosis. Hum Mol Genet. 2005, 14: 1795-1803. 10.1093/hmg/ddi186.
Tyson C, Harvard C, Locker R, Friedman JM, Langlois S, Lewis ME, Van Allen M, Somerville M, Arbour L, Clarke L, McGilivray B, Yong SL, Siegel-Bartel J, Rajcan-Separovic E: Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am J Med Genet A. 2005, 139: 173-185.
Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M: Strong association of de novo copy number mutations with autism. Science. 2007, 316: 445-449. 10.1126/science.1138659.
Moon HJ, Yim SV, Lee WK, Jeon YW, Kim YH, Ko YJ, Lee KS, Lee KH, Han SI, Rha HK: Identification of DNA copy-number aberrations by array-comparative genomic hybridization in patients with schizophrenia. Biochem Biophys Res Commun. 2006, 344: 531-539. 10.1016/j.bbrc.2006.03.156.
Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K: Large recurrent microdeletions associated with schizophrenia. Nature. 2008, 455: 232-236. 10.1038/nature07229.
Sutrala SR, Goossens D, Williams NM, Heyrman L, Adolfsson R, Norton N, Buckland PR, Del-Favero J: Gene copy number variation in schizophrenia. Schizophr Res. 2007, 96: 93-99. 10.1016/j.schres.2007.07.029.
Stone JL, O'Donovan MC, Gurling H, Kirov GK, Blackwood DH, Corvin A, Craddock NJ, Gill M, Hultman CM, Lichtenstein P, McQuillin A, Pato CN, Ruderfer DM, Owen MJ, St Clair D, Sullivan PF, Sklar P, Purcell SM, Stone JL, Ruderfer DM, Korn J, Kirov GK, Macgregor S, McQuillin A, Morris DW, O'Dushlaine CT, Daly MJ, Visscher PM, Holmans PA, O'Donovan MC, Sullivan PF, Sklar P, Purcell SM, Gurling H, Corvin A, Blackwood DH, Craddock NJ, Gill M, Hultman CM, Kirov GK, Lichtenstein P, McQuillin A, O'Donovan MC, Owen MJ, Pato CN, Purcell SM, Scolnick EM, St Clair D, Stone JL, Sullivan PF, Sklar P, O'Donovan MC, Kirov GK, Craddock NJ, Holmans PA, Williams NM, Georgieva L, Nikolov I, Norton N, Williams H, Toncheva D, Milanova V, Owen MJ, Hultman CM, Lichtenstein P, Thelander EF, Sullivan P, Morris DW, O'Dushlaine CT, Kenny E, Waddington JL, Gill M, Corvin A, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Puri V, Krasucki R, Lawrence J, Quested D, Bass N, Curtis D, Gurling H, Crombie C, Fraser G, Kwan SL, Walker N, St Clair D, Blackwood DH, Muir WJ, McGhee KA, Pickard B, Malloy P, Maclean AW, Van Beck M, Visscher PM, Macgregor S, Pato MT, Medeiros H, Middleton F, Carvalho C, Morley C, Fanous A, Conti D, Knowles JA, Ferreira CP, Macedo A, Azevedo MH, Pato CN, Stone JL, Ruderfer DM, Korn J, McCarroll SA, Daly M, Purcell SM, Sklar P, Purcell SM, Stone JL, Chambert K, Ruderfer DM, Korn J, McCarroll SA, Gates C, Daly MJ, Scolnick EM, Sklar P: Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008, 455: 178-179. 10.1038/nature07239.
Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J: Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008, 320: 539-543. 10.1126/science.1155174.
Wilson GM, Flibotte S, Chopra V, Melnyk BL, Honer WG, Holt RA: DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signalling. Hum Mol Genet. 2006, 15: 743-749. 10.1093/hmg/ddi489.
Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M: Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008, 40: 880-885. 10.1038/ng.162.
Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, Ge D, Heinzen EL, Maia JM, Shianna KV, Weale ME, Cherkas LF, Clement G, Spector TD, Gibson G, Goldstein DB: A Genome-wide Study of Common SNPs and CNVs in Cognitive Performance in the CANTAB battery. Hum Mol Genet. 2009
Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R: Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008, 17: 458-465. 10.1093/hmg/ddm323.
American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders. 1994, Washington, DC: American Psychiatric Press
Andreasen NC: Negative symptoms in schizophrenia. Definition and reliability. Arch Gen Psychiatry. 1982, 39: 784-788.
Andreasen NC: The scale for the assessment of positive symptoms (SAPS). 1984, Iowa: University of Iowa Press
Liu SK, Chiu CH, Chang CJ, Hwang TJ, Hwu HG, Chen WJ: Deficits in sustained attention in schizophrenia and affective disorders: stable versus state-dependent markers. Am J Psychiatry. 2002, 159: 975-982. 10.1176/appi.ajp.159.6.975.
Lin CC, Chen WJ, Yang HJ, Hsiao CK, Tien AY: Performance on the Wisconsin Card Sorting Test among adolescents in Taiwan: norms, factorial structure, and relation to schizotypy. J Clin Exp Neuropsychol. 2000, 22: 69-79. 10.1076/1380-3395(200002)22:1;1-8;FT069.
Lee CH, Fang CY, Sheu JJ, Chang Y, Takada K, Chen JY: Amplicons on chromosome 3 contain oncogenes induced by recurrent exposure to 12-O-tetradecanoylphorbol-13-acetate and sodium n-butyrate and Epstein-Barr virus reactivation in a nasopharyngeal carcinoma cell line. Cancer Genet Cytogenet. 2008, 185: 1-10. 10.1016/j.cancergencyto.2008.03.014.
Lee CH, Wu CC, Wu YN, Chiang HS: Gene copy number variations in Asian patients with congenital bilateral absence of the vas deferens. Hum Reprod. 2009, 24: 748-755. 10.1093/humrep/den413.
Database of Genomic Variants. [http://projects.tcag.ca/variation/]
Gene Expression Omnibus. [http://www.ncbi.nlm.nih.aov/geo/]
Bodin L, Beaune PH, Loriot MA: Determination of cytochrome P450 2D6 (CYP2D6) gene copy number by real-time quantitative PCR. J Biomed Biotechnol. 2005, 2005: 248-253. 10.1155/JBB.2005.248.
PubMed. [http://www.ncbi.nlm.nih.gov]
Kam M, Massare J, Gallinger S, Kinzie J, Weaver D, Dingell JD, Esufali S, Bapat B, Tobi M: Peutz-Jeghers syndrome diagnosed in a schizophrenic patient with a large deletion in the STK11 gene. Dig Dis Sci. 2006, 51: 1567-1570. 10.1007/s10620-006-9102-8.
Wallen-Mackenzie A, Mata de Urquiza A, Petersson S, Rodriguez FJ, Friling S, Wagner J, Ordentlich P, Lengqvist J, Heyman RA, Arenas E, Perlmann T: Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev. 2003, 17: 3036-3047. 10.1101/gad.276003.
Wang S, Sun CE, Walczak CA, Ziegle JS, Kipps BR, Goldin LR, Diehl SR: Evidence for a susceptibility locus for schizophrenia on chromosome 6pter-p22. Nat Genet. 1995, 10: 41-46. 10.1038/ng0595-41.
Goodman AB: Congenital anomalies in relatives of schizophrenic probands may indicate a retinoid pathology. Schizophr Res. 1996, 19: 163-170. 10.1016/0920-9964(96)88523-9.
Goodman AB: Three independent lines of evidence suggest retinoids as causal to schizophrenia. Proc Natl Acad Sci USA. 1998, 95: 7240-7244. 10.1073/pnas.95.13.7240.
Krezel W, Ghyselinck N, Samad TA, Dupe V, Kastner P, Borrelli E, Chambon P: Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. Science. 1998, 279: 863-867. 10.1126/science.279.5352.863.
Mena MA, Casarejos MJ, Bonin A, Ramos JA, Garcia Yebenes J: Effects of dibutyryl cyclic AMP and retinoic acid on the differentiation of dopamine neurons: prevention of cell death by dibutyryl cyclic AMP. J Neurochem. 1995, 65: 2612-2620.
Samad TA, Krezel W, Chambon P, Borrelli E: Regulation of dopaminergic pathways by retinoids: activation of the D2 receptor promoter by members of the retinoic acid receptor-retinoid × receptor family. Proc Natl Acad Sci USA. 1997, 94: 14349-14354. 10.1073/pnas.94.26.14349.
Acknowledgements
This study was supported by grants from the National Science Council, Taiwan (NSC91-2314-B-002-216; NSC91-3112-B-002-011; NSC 92-3112-B-002-019; NSC93-3112-B-002-012; NSC94-3112-B-002-020; NSC95-3112-B002-011; NSC 95-2314-B-002-313; NSC96-3112-B-002-011; and, NSC97-3112-B-002-046).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Author HHG designed the study and managed the literature searches. Author LCH designed the study, undertook the array CGH analysis, and wrote the draft of the manuscript. Author LCM recruited the participants, undertook the statistical analysis, and wrote the draft of the manuscript. Author WCC collected the samples. Author CSM performed the qPCR experiments. All authors contributed to and have approved the final manuscript.
Chia-Huei Lee, Chih-Min Liu contributed equally to this work.
Electronic supplementary material
12929_2009_116_MOESM2_ESM.PDF
Additional file 2: Supplementary table 2. Comparison of incidence of CNVs in schizophrenics and in control subjects. (PDF 9 KB)
12929_2009_116_MOESM3_ESM.DOC
Additional file 3: Supplementary figure 1. The quantitative real-time PCR (QPCR) results for potential candidate genes identified by array CGH. The fold change in gene copy number for each indicated target gene relative to the endogenous reference gene (ATP2B4) was compared for the genomic DNA samples from affected sib pairs with at least one showing positive results by array CGH. The fold change for each target gene and ATP2B4 of control sample was set at 1. The normalized fold changes were interpreted as follows: No change (0.7-1.4, white bar), homozygous loss (< 0.3, black bar), over representation (> 1.4, black bar) and ambiguous (0.3-0.7, gray bar). (DOC 166 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lee, CH., Liu, CM., Wen, CC. et al. Genetic copy number variants in sib pairs both affected with schizophrenia. J Biomed Sci 17, 2 (2010). https://doi.org/10.1186/1423-0127-17-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1423-0127-17-2