Abstract
Arrhythmia is a heart condition that poses a severe threat to life and requires prompt medical attention. One of the challenges in detecting arrhythmias accurately is that incorrect diagnoses can have severe consequences. In light of this, it is critical to develop a solution that is both effective and reliable. In this study, we propose a residual Convolution Neural Network Bidirectional Long Short-Term Memory (DeepResidualBiLSTM) model for classifying Arrhythmia types, which addresses the vanishing gradient problem and captures the relevant features in the signals’ long dependencies. The model is characterized by its simplicity, stability, and ability to extract meaningful features effectively. Using two well-known datasets, the experimental results demonstrate exceptional accuracy, precision, and recall values of approximately 99.4% at the early stage of 20 epoch training. Furthermore, the model demonstrates a remarkable ability to discriminate between Arrhythmia classes under varying thresholds using the ROC curve metric, with a high value, in most cases, of 100% for accurately detecting positive cases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Arrhythmia is a heart disorder indicated by abnormal electrical activity in the heart [6, 9]. This can manifest in the form of tachycardia, bradycardia, or irregular heartbeats. The causes of arrhythmia can vary from heart disease, high blood pressure, diabetes, genetics, certain medications, and alcohol or drug abuse [17]. There are several distinct types of arrhythmias, including atrial fibrillation, ventricular tachycardia, and bradycardia. Some arrhythmias are benign and require no treatment, while others can be life-threatening and require immediate medical attention [10]. However, arrhythmias can be detected by experts through the analysis of the electrical signals captured by an electrocardiogram (ECG), which is a diagnostic test that measures and records the heart’s electrical activity. The ECG is a valuable tool used by healthcare providers to assess heart health and develop appropriate treatment plans. The detection of arrhythmias can be a challenging problem, as it often requires the analysis of complex ECG signals [8, 29, 31]. Accordingly, the challenges associated with arrhythmia detection include [22, 25,26,27]: identifying rare arrhythmias of a short period; classifying different types of arrhythmias of unique characteristics; dealing with noise and artefacts of ECG signals; and specialists may vary in their levels of knowledge, experience, and ability to interpret ECG data. To address the challenges, we need advanced signal-processing techniques and machine-learning algorithms. In particular, the process of detecting arrhythmia begins with analyzing the ECG signals using various signal processing techniques to extract features and then being used as input for deep learning models.
Numerous applications, including image identification, audio recognition, natural language processing, and gaming, have extensively used deep learning [1,2,3,4]. Because the algorithms must discover patterns in the data to produce precise predictions, it necessitates a huge volume of tagged data. Therefore, improvements in processing power and a vast amount of labelled training data have enabled the creation of deep learning. Convolutional neural networks (CNNs) or recurrent neural networks (RNNs), however, can be utilized in deep learning to extract features from ECG signals. In the case of RNNs, the network takes into account the temporal dependencies in the signal and can learn to extract features such as rhythm and beat patterns. These features can then be used for tasks such as arrhythmia classification or heart disease prediction [7, 12, 14, 16, 19, 20, 23, 24, 28]. Regrading ECG signals, heartbeats can be classified into five types: non-ectopic (or normal beat (N)), supra ventricular ectopic (S), ventricular ectopic (V), fusion (F) and unknown beats (Q). There are various types of arrhythmias and each type is associated with a pattern, and as such, it is possible to identify and classify its type. Arrhythmias can be classified into two major categories. The first category consists of arrhythmias formed by a single irregular heartbeat, herein called morphological arrhythmia. The other category consists of arrhythmias. Arrhythmia can be detected by identifying an individual abnormal heartbeat, which can occur in isolation or sequentially. However, the early and accurate detection of heartbeat arrhythmia is crucial in the timely treatment and management of patients’ cases. Physicians would rely heavily on a clear diagnosis offered by the proposed model in predicting the complications and risks associated with the arrhythmia, a better prognosis that would assist the patients in their future course of action, and a basis for scientific research in the field of arrhythmia understanding and treatment design.
We propose a deep residual CNN bidirectional LSTM (DeepResidualBiLSTM) model that consists of two main components for extracting (or filtering) features from the input ECG signals. The first component employs the residual networks to mitigate the gradient vanishing issues in updating the weights of deep layers. This ensures that the gradient information can be effectively propagated through the neural network, leading to improvement in training performance. The second component exploits the bidirectional LSTM model that receives the features from the first component and further conserves the more relevant features during training. The model captures the dependencies in the ECG features in both forward and backward directions, which can result in a more robust and accurate representation of the signal. Furthermore, the DeepResidualBiLSTM composes a deep three-layer ANN with a soft-max function for classifying the input signal types. What differentiates the proposed approach are the simplicity, high performance, stability, and ease of understanding from an explainable AI systems perspective.
The organisation of the paper is as follows: Sect. 2 discusses the research of various approaches in detecting arrhythmia in ECG signals. Section 3 presents the proposed model architecture and the reason behind using the inherited components. We clarify the experiment settings and evaluate the proposed model using a validation technique in Sect. 4. In addition, the discussion of the results is presented on two common datasets. Section 5 summarizes the paper and opens the door for future work.
2 Related Works
Arrhythmia detection using deep learning involves training a model to recognize patterns in ECG data that indicate the presence of arrhythmias. This can be done using various techniques such as convolutional neural networks (CNNs), recurrent neural networks (RNNs), and long short-term memory (LSTM) networks. One common approach is using CNNs to classify ECG segments as normal or abnormal, based on their similarity to known arrhythmia patterns. Research in this field has shown promising results, with deep learning models achieving high accuracy in detecting arrhythmias such as atrial fibrillation and ventricular tachycardia. Table 1 shows a summary of the following discussed research works.
Several studies employed different ML approaches to classify heartbeat arrhythmia. Kachuee et al. [15] proposed a model consisting of 13 layers of 1D CNN with the aim of predicting 5 types of arrhythmias following the AAMI EC57 standard. To assess the effectiveness of their approach, they conducted evaluations using PhysionNet’s MIT-BIH and PTB Diagnostics datasets. The results revealed that their proposed method achieved average accuracies of 93.4% and 95.9% for arrhythmia classification and MI classification, respectively.
The study in [12] proposed a DenseNet transfer learning model and GRU network with attention (DenseNet-attention-GRU) architecture to tackle the inter-patient ECG classification issue. They evaluated the proposed architecture using two datasets focused on classifying the supraventricular (S) and ventricular (V) detection, which obtained F1-score results of 61.25 (20% improvement) for S detection and 89.75 (1.31% improvement) for V detection, respectively.
In their study, Xu et al. [30] introduced a combined network comprising the Squeeze-and-Excitation Network (SENet) and a biLSTM model for the purpose of diagnosing heart arrhythmias. The network exhibited remarkable performance, achieving a recognition sensitivity of 95.90%, accuracy of 95.90%, and specificity of 96.34%.
Ullah et al. [28] proposed three residual deep learning techniques: CNN, CNN+LSTM, and CNN+LSTM+Attention models for detecting arrhythmias. The CNN is used for automatically extracting features, while the LSTM model is used to learn the dependency between features for the classification part. The accuracy of the models is 99.3%, 99.3%, and 99.29%, respectively.
Jamil and Rahman [14] suggested a framework of four blocks that comprise the pipeline for classifying the arrhythmia. The blocks prepare the ECG signals by segmenting them into beats and then converting the beats into a 2D time-frequency representation using the continuous wavelet transform (CWT). A proposed CNN model with attention block (or ArrhythmiaNet) is used to extract local and global features. Then, the features are processed into the third block for further reduction using the k-means clustering technique. The last block is the support vector machine (SVM) classifier model for detecting the 17 classes of heartbeat arrhythmia categories. The proposed framework showed superior results of 99.84% accuracy with an SVM classifier, a sensitivity value of 100%, and an F1-score of 99%. The classification accuracy of ArrhythmiaNet with a kNN classifier was 98.64%, which is insufficient compared to an SVM classifier.
Liu et al. [19] proposed an auto-encoder deep learning network to extract the time series of ECG signals. The network consists of encoding and decoding components, each of which comprises stacked layers of the LSTM model. The classification part of the proposed model receives as input the compressed features in the bottleneck between the encoder and decoder to detect the arrhythmia classes as an output target. However, the network tackles the problem of vanishing gradient descent due to its simplicity by not using the complexity of CNN structures for updating the network weights and bias. The experiment results showed that the proposed model achieved 98.57% accuracy, 97.98% recall and 97.55% positive precision.
Qin et al. [23] concentrated on the problem of imbalanced ECG dataset distribution. They proposed a framework of two architectures to handle the issue and perform classification. The improved WGAN-GP (Wasserstein Generative Adversarial Networks-Gradient Penalty) is a pre-processing model of one-dimensional convolutional layers in the generator and discriminator components of the WGAN network. The improvement part of WGAN-GP is the Bi-directional gate recurrent unit (Bi-GRU) layer on the generator for updating the parameter values to generate ECG time series by learning the distribution of the real data. The prepared dataset is then fed into the SE-ResNet1D CNN network to classify the ECG signals. The SE-ResNet1D is a one-dimensional deep residual network (ResNet) composed of squeeze block and incentive block structures on top of the ResNet model. The proposed WGAN-GP+SEResNet1D method yields an acceptable precision of 95.8%, recall of 96.75%, and F1 of 96.27% on the balanced dataset.
Rahul and Sharma [24] proposed a five-stage methodology for automatically classifying cardiac arrhythmias, which included pre-processing, segmentation, normalization, 1D CNN, and BiLSTM. The pre-processing pipeline removes noise from the input raw ECG signal using stationary wavelet transform (SWT) for decomposition, a two-stage median filter, and the Savitzky–Golay filter and then reconstructs the signal using an inverse SWT (ISWT) filter. Due to long ECG records, the signals are segmented into 5-second segments during the segmentation process, followed by normalization to minimize data redundancy. The 1D-CNN model extracts the relevant features, which are then sent into the BiLSTM classifier. The experiments showed a superior accuracy of 99.41% achieved using 10-fold cross-validation. By a simple data preparation, [13] proposed a straightforward 1D CNN Bi-LSTM model for detecting five distinct types of ECG signals. The model achieves promising performance results of 98%, 90.96%, and 91% accuracy, specificity, and recall using the MIT-BIH dataset.
Liu et al. [20] proposed an ECG detection system with three components: pre-processing, feature extraction, and classification. The pre-processing process has three phases: re-sampling, segmentation, and max-min normalization. The feature extraction process uses the ECG-Convolution-Vision Transformer Network (ECVT-Net) model to extract features from ECGs. The 1D CNN block extracts high-dimensional abstract features, while the transition block in the system prepares the extracted features to be employed in the ViT transformer. Six ViT transformer blocks learn the dependencies between features in long-term sequences. The system also comprises a fully connected multi-layer perceptron (MLP) and soft-max layers for the binary classification of target outputs. The conducted experiments yielded stable results with different levels of noise interference and showed accuracy, precision, and sensitivity values of 98.88%, 98.84%, and 98.94%, respectively.
Li et al. [18] proposed an arrhythmia classification structure. The structure incorporates overlapping segmentation of the ECG signal and majority voting to re-label each 5-second signal segment to balance the MIT-BIH dataset. The Daubechies DWT denoises segments and Z-score standardization is performed. The classification and extraction part is an improved 1D CNN residual network, which is several convolution kernels of various sizes with a focal loss function to improve the perception of different scales in the convolution layer and increase the diversity of features. The experiment results give 94.54%, 35.22%, and 88.35% sensitivity values for normal, supraventricular ectopic, and ventricular ectopic segments, respectively.
Kim et al. [16] proposed a data augmentation framework, a stacked 1D CNN residual network with a squeeze and excitation block, and a BiLTSM model. The SMOTE augmentation method is applied as a pre-processing step to tackle the problem of imbalanced data by oversampling the minority classes. The 1D CNN and BiLSTM extract the global features from balanced raw ECG. Then, the global feature vector is input into a fully connected ANN of two dense layers and two activation layers for arrhythmia classification. Using the 10-fold cross-validation technique, the experimental results showed an accuracy value of 99.2% and an F1-score value of 91.69% on the MIT-BIH dataset.
Unlike previous studies that suggested using large networks for small input signals, this research aims to explore different aspects of heartbeat arrhythmia using a compact and efficient model. The hybrid model proposed in this study combines the residual network, BiLSTM model, and fully connected ANN layer to leverage their strengths. Furthermore, new layers have been introduced to improve prediction accuracy. As a result, the proposed model is comprehensive, lightweight, and efficient, making it suitable for various types of heartbeat arrhythmia without the need for extra resources or effort.
3 Deep Residual CNN Bidirectional LSTM Model
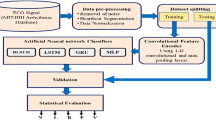
The proposed DeepResidualBiLSTM model comprises two main components: the residual transfer learning model and the sequence long memory in the bidirectional LSTM model. The heartbeat signal is a collection of data that contains numerous hidden indications that can be used to detect arrhythmia problems. This section covers the proposed methodology and the rationale behind using this structure to classify arrhythmia issues. The design is depicted in Fig. 1, demonstrating the input signals, a residual transfer learning model for feature extraction, a bidirectional LSTM model that filters features based on dependencies, and a deep three-layered ANN with a soft-max function for classifying the input signal types.
3.1 Residual Deep Transfer Learning Model for Feature Extraction
The residual learning models handle the problem of vanishing gradients while updating the network weights of long layers [5]. The vanishing gradient leads to a slow gradient descent with zero or small values at the beginning of the network. This is insignificant in terms of optimizing layer parameters and adding more layers to the network. The residual networks skip a set of layers to propagate the large loss values backwards during the training. Hence, the residual network uses skip connections as a core building block to tackle the problem of gradient vanishing and improve the performance accuracy of the network by extracting more pertinent features from the raw inputs. Figure 2 shows mathematically the architecture of the residual block. The output x from the previous layer is combined with the output of a specific successor stacked layers g(x); \(f(x) = g(x) + x\).
The skip connection is a shortcut that transfers the output of the previous layer to the output of stacked layers of the Conv1D and ReLU processes. The skip connection technique in the proposed model is shown at the beginning and three times after the ReLU process, in which the loss values are kept up-to-date in a limited number of layers to minimize the performance error and overfitting problem. Furthermore, we apply L2 regularization, often referred to as weight decay, to every parameter within the convolutional layers during optimization. The L2 regularization, a prevalent method applied in CNNs, aids in averting overfitting by imposing penalties on significant weights or inducing sparsity. In the framework of a 1D convolutional layer, the regularization term is integrated into the loss function. The standard 1D convolutional layer equation is:
where \(z_i\) is the output at position i, \(x_{i+j}\) is the input at position \(i+j\), \(w_j\) is the weight at position j, b is the bias term and k is the size of the convolutional kernel.
With L2 regularization, the loss function becomes:
where N is the number of samples, \(y_i\) is the true output at position i, \(\lambda \) is the regularization parameter.
The first term in the loss function is the standard mean squared error, and the second term is the L2 regularization term. The regularization term penalizes large values of the weights, helping to keep them small and preventing overfitting. Concerning data augmentation, we refrained from its application within the specific context of managing heartbeat signals. This approach may entail certain drawbacks, such as: (1) The risk of overfitting arises with aggressive data augmentation, particularly when the augmented data closely resembles the original, leading to a potential lack of generalization, (2) Data augmentation has the propensity to introduce unrealistic or semantically inconsistent samples, thereby posing challenges to the fidelity of the augmented dataset and (3) the quality of augmented data is subject to variation. Certain transformations may inadvertently distort the original information, compromising the reliability of the augmented samples.
The distinguishing feature of the proposed model in comparison to previous works is the adaptation of the number of layers to the size of input signals. Since the input signals are not wide enough to accommodate numerous layers, we conducted experiments to identify the optimal number of layers for an efficient model structure.
3.2 Bidirectional LSTM Model for Long Sequence Learning
The proposed approach uses BiLSTM as a base layer between the CNN model and fully connected dense layers. The BiLSTM model preserves the flow of information by allowing it to flow both forward (from the past to the future) and backward (from the future to the past) [11]. The BiLSTM model’s essential component, which is the LSTM model, uses an activation function to organise the input flow and converge it at the same level. The LSTM layers are a series of layers that are iteratively applied to preserve the feature maps acquired from the output of the CNN model as input sequence features [21]. The LSTM model is one of the Recurrent Neural Network (RNN) models that comprise a chain of periodic modules in the form of neural networks. The connections for forwarding information in RNN models depend on the time steps; thus, the output at the one-time step becomes the input at the following time step. The model precisely considers what is persisting from the preceding elements at each element of the input sequence. This phenomenon avoids the issue of long-term dependency. As a result, long-short-term memory (LSTM) is the most common cell in the architecture of LSTM models, which is expressed in a single cell that is connected in time. This architecture is represented in Fig. 3. This preserves a cell state and a carry to ensure that the gradient-based information is not lost throughout the processing of the sequence.
As shown in Fig. 3, the hidden state H(t) value of the cell that is updated over time, and the cell state C(t) handles memory over the long term, are the two influential values making up the LSTM model. The top line of the LSTM cell adjusts the cell state by adding or removing information where the function of each cell element is determined by the parameters (weights) learned during training. The cell elements of the LSTM models are forget, input, and output gates. The forget gate F(t) adjusts the connection of the input X(t) and the previous hidden state \(H(t - 1)\) to the cell state C(t). This allows the cell to retain or forget X(t) where the \(H(t-1)\) depends on the binary output of the Sigmoid functions. The input gate I(t) and \(\underline{I}(t)\) decide whether to feed the input value to the cell state C(t). The output gate is used for producing predictions at each time step. The output gate O(t) determines the final value based on the cell state C(t). This process is formulated in a set of equations shown in Eqs. 3–8. There are four major functions used in LSTM models: sigmoid (\(\sigma \)), hyperbolic tangent (tanh), multiplication (\(\times \)), and sum (\(+\)). These functions make the processes easier to update the weights during the back-propagation process.
where W and B denote the weight matrix and bias vector, respectively. The sigmoid function is denoted by sigma(cdot), and the hyperbolic tangent function is denoted by tanh(cdot). The weights for the forward, input, and output gates are denoted by \(W_f\), \(W_i\), and \(W_o\), respectively. The use of LSTM layers is motivated by their robustness in recognizing sequence signals without vanishing the gradient. The vanishing gradient shows a slight decrease in the values of the weights, which appear to have not changed during the training process. The back-propagation gradient descent updates the weights of whole networks, including the LSTM layers’ weights. The BiLSTM, in the proposed model, comprises three parts, each of which has two LSTM models running in forward and backward directions. Two LSTM models connect the input and the activation function for the output. However, the BiLSTM more effectively captures contextual information, provides reliable reverse evaluation, and thereby facilitates robust cross-validation of the results. The integration of BiLSTM markedly amplifies the predictive power of a machine learning model. This enhancement arises from training the model with values from both forward and backward directions, consequently elevating the model’s learning rate substantially. We use two BiLSTM models before fully connected networks to handle dependencies among the features from the residual network and to avoid the long-term dependency problem. These inter-dependencies form the context features to enhance the proposed model’s relevancy.
4 Experimental Settings and Results
4.1 Datasets Settings
We evaluate the proposed ResidualBiLSTM model using two widely used heartbeat classification datasets: the modified MIT-BIH Arrhythmia datasetFootnote 1 [15] and the PTBDB diagnostic ECG datasetFootnote 2 with a sampling rate of 125 Hz. As a preprocessing measure, a band-pass filter is used to eliminate undesirable noise or interference by permitting only specific frequency ranges to pass through while suppressing those beyond it. The signal is then partitioned into smaller segments, each consisting of a single heartbeat, and any segments that contain artifacts such as electrode noise or muscle movement are removed. Additionally, to ensure consistency, every sample is resized to a fixed dimension of 188 by cropping, down-sampling, and padding with zeroes. The MIT-BIH dataset includes five main categories of 109,446 beats: Non-ecotic beats (normal beat (N)), Supraventricular ectopic beats (S), Ventricular ectopic beats (V), Fusion Beats (F), and Unknown Beats (Q) with 90,589, 8039, 7236, 2779, and 803 beats, respectively. The PTBDB dataset consists of two classes: normal and abnormal.
Figure 4 depicts the five types of beats. The normal beats originate from the sinoatrial (SA) node and travel to the atria and then to the ventricles. On the other hand, supraventricular ectopic beats are extra heartbeats that start in the atria or another part of the atrioventricular (AV) node. The ventricular ectopic beats originating from the ventricles, the lower chambers of the heart, lead to a premature contraction of the heart muscle. A fusion beat is a type of abnormal heartbeat that occurs when two electrical impulses are present in the heart simultaneously, one from the normal pacemaker in the heart (the sinoatrial, or SA, node) and one from another location in the heart, leading to a combination or hybrid beat as the heart muscle contracts partially with the normal beat and partially with the ectopic beat.
4.2 Evaluation and Validation Settings
To evaluate the classifier performance, we employ an infrastructure known as KerasFootnote 3 2.14.0, a Python-based deep learning API built on the TensorFlow machine learning platform. The experiments are executed on the KaggleFootnote 4 framework using GPU P100 with a training time of 334.4 s. In terms of evaluation metrics, We use five evaluation metrics; accuracy, precision, recall, f1-score and Receiver Operating Characteristic (ROC) Under-curve. We discuss the Roc under-curve metric in the experiment results and discussion section.
-
Accuracy: is calculated as the ratio of correct predictions made by a model to the total number of predictions. Essentially, it reflects the proportion of accurate predictions made by the model. However, in certain situations where the majority class dominates, such as imbalanced datasets, accuracy can be misleading. For example, a model that consistently predicts the majority class can still have high accuracy, even if it does not make any correct predictions for the minority class. Other metrics like precision, recall, F1-score, or the AUC-ROC are frequently employed to obtain a more comprehensive evaluation of a model’s performance in such cases.
$$\begin{aligned} \textrm{Accuracy} = \frac{TP + \textrm{True Negative}~(\textrm{TN})}{\textrm{TP}+\textrm{FP}+\textrm{FN}+\textrm{TN}} \end{aligned}$$(9) -
Precision: measures the fraction of positive predictions that are actually correct, i.e., the ratio of true positive predictions to the sum of true positive and false positive predictions. Precision is particularly useful when the cost of false positives is high.
$$\begin{aligned} \textrm{Precision} = \frac{\textrm{True Positive} ~(\textrm{TP})}{\textrm{TP} + \mathrm{False Positive~(FP)}} \end{aligned}$$(10) -
Recall: (also known as Sensitivity or True Positive Rate) measures the fraction of positive cases that are correctly identified by the classifier, i.e., the ratio of true positive predictions to the sum of true positive and false negative predictions. The recall is particularly useful when the cost of false negatives is high.
$$\begin{aligned} \textrm{Recall} = \frac{\textrm{TP}}{\textrm{TP} + \mathrm{False Negative~(FN)}} \end{aligned}$$(11) -
F1-score: is a harmonic mean of precision and recall, and provides a balance between the two metrics. The F1-score has a score of 1 represents a perfect balance between precision and recall and a score closer to 0 indicates a poor balance. The F1-score is useful in cases where a balance between precision and recall is desired, such as in imbalanced datasets where there is a need to minimize false negatives.
$$\begin{aligned} F1\text{- }score = 2 \times \frac{\textrm{Precision} \times \textrm{Recall}}{\textrm{Precision} + \textrm{Recal}l} \end{aligned}$$(12) -
ROC Under-curve: serves as a graphical representation for evaluating the performance of a model. It visually depicts the trade-off between the true positive rate (sensitivity) and the false positive rate (1-specificity) under various threshold settings. The False Positive Rate (1-Specificity) is the ratio of incorrectly predicted negative instances to the total actual negative instances.
$$\begin{aligned} \mathrm{False~Positive~Rate~(1-Specificity)} = \frac{\textrm{FP}}{\textrm{FP}+ \textrm{TN}} \end{aligned}$$(13)The ROC curve provides a comprehensive evaluation of performance across diverse threshold settings, proving especially pertinent in scenarios characterized by imbalanced class distributions, as observed in utilized datasets, notably within the realm of medical diagnostics. Furthermore, the ROC AUC Curve succinctly encapsulates the classifier’s performance in a singular scalar value, furnishing a condensed measure of its discriminatory efficacy. Consequently, it emerges as a convenient and widely endorsed metric for comparative assessments, particularly in medical applications where precision and sensitivity hold paramount significance.
Table 2 shows that a batch size of 32 was utilized for the datasets as a moderated value to balance between the robustness of stochastic gradient descent and the efficiency of batch gradient descent due to limited resources. The initial learning rate was set to 0.001 and was adjusted using a decay strategy, where the interval of decay was 16 and the rate was reduced by a factor of 0.5 after each interval of 5 epochs (or patience), contingent upon the absence of improvement in validation accuracy. This approach stabilized the results and improved the network’s performance. The Adam optimizer was used to update the weights with its parameters as shown. The Adam optimizer is considered one of the best adaptive optimizers in most research cases. It contributes to the benefits of Adadelta and RMSprop by incorporating the retention of an exponentially decaying average of previous gradients, akin to the concept of momentum. The convolutional kernel size and the number of BiLSTM units were optimized experimentally where the best performance was achieved with a kernel size of 6 and 64 BiLSTM units, respectively. The model undergoes training for 100 epochs, despite the stabilization of both training and validation accuracy at epoch 30. This decision is made to ensure the scalability of the training model across a substantial number of epochs while maintaining stable accuracy from the early stages.
To mitigate the possibility of certain instances not participating in both the training and validation stages, we adopt a stratified 10-fold cross-validation technique during the evaluation phase. This approach involves iteratively selecting 10% of the dataset for validation (or testing) and 90% of the dataset for training (comprising the remaining 9 folds), and repeating this procedure for all instances. Consequently, each instance takes part in both the training and testing (or validation) stages ten times. We create a classifier model and assess its performance in each procedure. Then, a representation of the overall average performance is shown. By employing such a method, we make sure that the complete dataset is used during the training and testing stages, lowering the possibility of over-fitting.
4.3 Experiment Results and Discussion
The training phase constructs the model in an iterative process to reduce the error between the actual and predicted targets. During the training phase, the model might tightly fit the training set and performs worse in testing sets, leading to an overfitting problem. A validation set of the dataset is used to adjust the hyper-parameters during training and to tackle overfitting. Thus, we split the datasets to generate a validation set of 10% in comparison to the training and test sets. Figure 5 shows the training accuracy and error loss of the proposed model on two datasets. The x-axes represent the number of iterations for each input batch (or epoch). The y-axes represent the model accuracy or error loss on the training and validation sets.
As illustrated in Fig. 5, the accuracy of training and validation sets are similar, with a discrepancy of up to 2% in the MIT-BIT dataset and 0.08% in the PTBDB dataset. This indicates the proposed model’s robustness and ability to accurately predict arrhythmia problems in training and unseen validation sets. The accuracy reached 98.9% at 20 epochs, steadily increasing to 100% ensuring the high accuracy performance at an early stage. This reduces the training time selecting smaller epoch values when we decide to increase the dataset for updating the model offline. Furthermore, the low loss error of less than or equal to 1% at epoch 20 ensures that the model has good performance and minimal over-fitting for the PTBDB dataset.
The training phase clarifies the effectiveness of the proposed model and stability during training at epochs 20; the reason is due to using skip-connection to update the weights in the feature extraction and the BiLSTM approach to maintain the relevant features in long sequences. The training phase also reduces the time complexity in which the experiment setting is better calibrated at early stages. The results show the model’s efficacy by minor differences between training and validation accuracy and loss values.
The proposed model was evaluated during the test phase using 10-fold cross-validation. Table 3 displays the results of the evaluation using precision, recall, f1-score, and accuracy on two datasets. The experimental findings reveal that our proposed model achieves higher precision, recall, and f1-score values of approximately 97% or above in the N, V, and Q classes. Conversely, there is only a minor improvement observed in the S and F classes. When compared to the research in Table 1, our model improves upon previous results. For example, in [12], the f1-score for S and V classes was 61.25% and 89.75%, respectively, while the proposed model improved upon these values by 28% and 6%, respectively. Additionally, [18] showed recall values of 94.54%, 35.22%, and 88.35% for the N, S, and V classes, respectively, while the proposed model improved upon these values by 5%, 55.4%, and 7%, respectively. Although prior research used sampling techniques to tackle imbalanced data and attained evaluation metric values of around 99%, the proposed model for the accuracy, recall (sensitivity), and F1-scores achieved promising results with approximately 1%\(-\)1.5% improvements.
To provide an evaluation and comparison perspective, we particularly choose the hybrid models as in Table 4. These models have been assessed using the same dataset and number of classes as our proposed model. When comparing our proposed model to related works, we observe remarkable accuracy results and comparable recall outcomes across all classes on average. What sets our proposed model apart from other related works is its reliance on the original data, yet it continues to perform exceptionally well, demonstrating its robustness. This signifies that our proposed model outperforms previous methods in terms of class types, overall accuracy, and recall values.
We evaluate the proposed model in an in-depth manner using the ROC (Receiver Operating Characteristic) Under-curve area as depicted in Fig. 6. The ROC curve is a graphical representation of the performance of a classifier system as the discrimination threshold is varied. The area under the ROC curve (AUC) measures the ability of the classifier to discriminate between positive and negative classes, which runs from 0.5 to 1, with 1 denoting a perfect classifier and 0.5 denoting a classifier with no discriminating ability. As shown in Fig. 6, the True Positive Rate (TPR), also known as Recall, is plotted on the y axis of the ROC curve, while the False Positive Rate (FPR) is plotted on the x axis. False Positive Rate (FPR), also known as fall-out, is the product of the number of false positives divided by the total of false positives and true negatives. On the other hand, recall is calculated by dividing the total number of true positives by the sum of true positives and false negatives as discussed before. The ROC curve visualizes the trade-off between these two metrics for different classification thresholds. A low fall-out (FPR) suggests a low false positive rate, indicating few false positive cases, whereas a high recall (TPR) shows a low false negative rate, indicating that most positive cases are accurately detected.
The ROC curve values for the five classes in the MIT-BIT dataset are displayed in Fig. 5a. The classes N, V, F, and Q demonstrate outstanding results with high recall values (i.e. 1) and low fall-out values (i.e. 0), signifying the reliability of the proposed model in classification. The S class achieved an exceptional area of 0.98. Meanwhile, the two classes in the PTBDB dataset display substantial results with an area under the curve of 1. The results ensure the robustness of the proposed model in discriminating the classes under varied thresholds with high recall values by detecting the most positive cases accurately.
The validation set results during the training phase demonstrate the ease of implementing our proposed model, a promising technology. Specifically, even in the early stages of 20 epochs, the model showed high accuracy and low error rates in classifying the arrhythmia problem, indicating the model’s strength in this field. Even in the testing phase using data with hidden labels, the proposed model obtains superior results on distinct metrics with encouraging values reaching up to approximately 99.4%. Hence, the advantages of the model architecture, in addition, to handling the problem of the vanishing gradient, are simplicity, high performance, stability, and ease of understanding.
To summarize, the proposed model outperforms previous works in terms of evaluation metrics for both validation and test sets, as evidenced by two well-known datasets. The model’s lightweight architecture, combined with its superior performance, establishes it as a promising technology for arrhythmia classification.
5 Conclusion and Future Work
Arrhythmia is a cardiac issue that poses a significant risk and threat to human life and it necessitates immediate medical intervention or treatment. The raw signals of ECG can provide indications for recognising issues based on the signal patterns. However, experts encounter challenges due to rare arrhythmias in a short period, exhibiting distinctive features of the signals with noise, and different specialists may interpret the signals in varying ways. To handle the challenges, this work aimed to propose an effective and reliable technique that automatically extracts the pertinent features from ECG signals and accurately classifies all forms of Arrhythmia. The proposed technique comprises three main components: a residual network, a BiLSTM model, and a fully connected ANN layer. Residual networks are employed to tackle the challenge of long distances for training network layers and updating weights for extracting low-dimensional features. Additionally, the BiLSTM network is utilized to learn the extended sequences of signal characteristics in both backward and forward directions. The final component, which involves the use of the softmax function, classifies various types of arrhythmia. Apart from the technique’s straightforwardness and efficiency, the experiments revealed outstanding results with accuracy, precision, and recall metrics reaching approximately 99.4%. These remarkable results are also demonstrated under different thresholds using the ROC metric.
In terms of future work, there are several avenues to explore for this model. One potential area is the expansion of the model’s capabilities to encompass additional types of heartbeat arrhythmia. Additionally, it would be beneficial to apply the proposed model to various datasets on heartbeat arrhythmia, which could be created and released for this purpose. Furthermore, the model could benefit from the implementation of alternative deep learning techniques to investigate any disparities in performance between the current model and any improved prediction models. These pursuits should be reserved for future research.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Change history
21 February 2024
A Correction to this paper has been published: https://doi.org/10.1007/s44196-024-00437-4
References
Alkhawaldeh, R.S.: Dgr: gender recognition of human speech using one-dimensional conventional neural network. Sci. Program. 2019 (2019)
Alkhawaldeh, R.S., Al-Dabet, S.: Unified framework model for detecting and organizing medical cancerous images in iomt systems. Multimed. Tools Appl. 1–28 (2023). https://doi.org/10.1007/s11042-023-16883-9
Alkhawaldeh, R.S., Khawaldeh, S., Pervaiz, U., Alawida, M., Alkhawaldeh, H.: Niml: non-intrusive machine learning-based speech quality prediction on voip networks. IET Commun. 13(16), 2609–2616 (2019)
Alkhawaldeh, R.S., Alawida, M., Alshdaifat, N.F.F., Alma’aitah, W., Almasri, A.: Ensemble deep transfer learning model for arabic (indian) handwritten digit recognition. Neural Comput. Appl. 1–15 (2021)
Amelio, A., Bonifazi, G., Cauteruccio, F., Corradini, E., Marchetti, M., Ursino, D., Virgili, L.: Representation and compression of residual neural networks through a multilayer network based approach. Expert Syst. Appl. 215, 119391 (2023)
Arifin, J., Sardjono, T.A., Kusuma, H.: Deep learning-based approaches for ecg signal arrhythmia: A comprehensive review. In: 2023 International Seminar on Intelligent Technology and Its Applications (ISITIA), pp. 417–421 (2023). https://doi.org/10.1109/ISITIA59021.2023.10221043
Arora, A., Taneja, A., Hemanth, J.: Heart arrhythmia detection and classification: A comparative study using deep learning models. Iran. J. Sci. Technol. Trans. Elect. Eng. (2023). https://doi.org/10.1007/s40998-023-00633-6
Corrado, C., Roney, C.H., Razeghi, O., Lemus, J.A.S., Coveney, S., Sim, I., Williams, S.E., O’Neill, M.D., Wilkinson, R.D., Clayton, R.H., et al.: Quantifying the impact of shape uncertainty on predicted arrhythmias. Comput. Biol. Med. 106528 (2023)
Dawood, M.: Cardiomyopathies, pp. 131–139. Springer International Publishing, Cham (2023). https://doi.org/10.1007/978-3-031-23062-2_17
Fawzy, A.M., Rivera-Caravaca, J.M., Underhill, P., Fauchier, L., Lip, G.Y.: Incident heart failure, arrhythmias and cardiovascular outcomes with sodium-glucose cotransporter 2 (sglt2) inhibitor use in patients with diabetes: Insights from a global federated electronic medical record database. Diab. Obes. Met. 25(2), 602–610 (2023)
Febrian, R., Halim, B.M., Christina, M., Ramdhan, D., Chowanda, A.: Facial expression recognition using bidirectional lstm-cnn. Proc. Comput. Sci. 216, 39–47 (2023)
Guo, L., Sim, G., Matuszewski, B.: Inter-patient ecg classification with convolutional and recurrent neural networks. Biocybern. Biomed. Eng. 39(3), 868–879 (2019)
Hassan, S.U., Mohd Zahid, M.S., Abdullah, T.A., Husain, K.: Classification of cardiac arrhythmia using a convolutional neural network and bi-directional long short-term memory. Digital Health 8, 20552076221102770 (2022)
Jamil, S., Rahman, M.: A novel deep-learning-based framework for the classification of cardiac arrhythmia. J. Imaging 8(3), 70 (2022)
Kachuee, M., Fazeli, S., Sarrafzadeh, M.: Ecg heartbeat classification: a deep transferable representation. In: 2018 IEEE International Conference on Healthcare Informatics (ICHI), pp. 443–444 (2018). https://doi.org/10.1109/ICHI.2018.00092
Kim, Y.K., Lee, M., Song, H.S., Lee, S.W.: Automatic cardiac arrhythmia classification using residual network combined with long short-term memory. IEEE Trans. Instrum. Measure. 71, 1–17 (2022)
Kloner, R.A.: Marijuana and electronic cigarettes on cardiac arrhythmias. Heart Rhy. 20(1), 87–88 (2023)
Li, Y., Qian, R., Li, K.: Inter-patient arrhythmia classification with improved deep residual convolutional neural network. Comput. Methods Prog. Biomed. 214, 106582 (2022)
Liu, P., Sun, X., Han, Y., He, Z., Zhang, W., Wu, C.: Arrhythmia classification of lstm autoencoder based on time series anomaly detection. Biomed. Signal Process. Control 71, 103228 (2022)
Liu, T., Si, Y., Yang, W., Huang, J., Yu, Y., Zhang, G., Zhou, R.: Inter-patient congestive heart failure detection using ecg-convolution-vision transformer network. Sensors 22(9), 3283 (2022)
Misgar, M.M., Mushtaq, F., Khurana, S.S., Kumar, M.: Recognition of offline handwritten urdu characters using rnn and lstm models. Multimed. Tools Appl. 82(2), 2053–2076 (2023)
Park, J., Lee, K., Park, N., You, S.C., Ko, J.: Self-attention lstm-fcn model for arrhythmia classification and uncertainty assessment. Artif. Intell. Med. 142, 102570 (2023). https://doi.org/10.1016/j.artmed.2023.102570
Qin, J., Gao, F., Wang, Z., Liu, L., Ji, C.: Arrhythmia detection based on wgan-gp and se-resnet1d. Electronics 11(21), 3427 (2022)
Rahul, J., Sharma, L.D.: Automatic cardiac arrhythmia classification based on hybrid 1-d cnn and bi-lstm model. Biocybern. Biomed. Eng. 42(1), 312–324 (2022)
Saito, K.: Potential and future challenges for cheyne-stokes breathing telemonitoring from continuous positive airway pressure devices. J. Clin. Sleep Med. JCSM 10456 (2023)
Shaik, T., Tao, X., Higgins, N., Li, L., Gururajan, R., Zhou, X., Acharya, U.R.: Remote patient monitoring using artificial intelligence: current state, applications, and challenges. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery, p. e1485 (2023)
Syed, T., Patel, N.R.: How can atrial fibrillation be detected and treated effectively? Trends Urol. Men Health 14(1), 5–10 (2023)
Ullah, W., Siddique, I., Zulqarnain, R.M., Alam, M.M., Ahmad, I., Raza, U.A.: Classification of arrhythmia in heartbeat detection using deep learning. Comput. Intell. Neurosci. 2021 (2021)
Wang, Y., Yang, G., Li, S., Li, Y., He, L., Liu, D.: Arrhythmia classification algorithm based on multi-head self-attention mechanism. Biomed. Signal Process. Control 79, 104206 (2023). https://doi.org/10.1016/j.bspc.2022.104206
Xu, X., Jeong, S., Li, J.: Interpretation of electrocardiogram (ecg) rhythm by combined cnn and bilstm. IEEE Access 8, 125380–125388 (2020). https://doi.org/10.1109/ACCESS.2020.3006707
Yesudasu, A.R.R., Revathi, N.N.S.P., Durga Prasad, P.R.L., Pujitha. K., Prabha, K.V.R.: A review on analysis of cardiac arrhythmia from heart beat classification. In: 2023 Second International Conference on Electronics and Renewable Systems (ICEARS), pp. 1464–1471 (2023). https://doi.org/10.1109/ICEARS56392.2023.10085295
Acknowledgements
This work was supported by Princess Nourah bint Abdulrahman University Researchers project number (PNURSP2024R387), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding
This work is funded.
Author information
Authors and Affiliations
Contributions
Conceptualization, AR, AS; methodology AR; software, AR, AB, GN; formal analysis, AR, AS, KA, GN; investigation AR, AS, AB, KA, GN; resources, AR, AE, AG, AM; data curation, AR, KA, GN; writing—original draft preparation, AR, AS; writing—review and editing, AR, AB, KA, GN, AE; visualization AR, AS, AG, AM; supervision, AR, A S, KA, AE; project administration, AR., GN; funding acquisition, KA, AG, AM, AR. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare(s) that there is no conflict of interest regarding the publication of this paper.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alkhawaldeh, R.S., Al-Ahmad, B., Ksibi, A. et al. Convolution Neural Network Bidirectional Long Short-Term Memory for Heartbeat Arrhythmia Classification. Int J Comput Intell Syst 16, 197 (2023). https://doi.org/10.1007/s44196-023-00374-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-023-00374-8