Abstract
Autophagy plays an active anti-viral role in plants. Increasing evidence suggests that viruses can inhibit or manipulate autophagy, thereby winning the arms race between plants and viruses. Here, we demonstrate that overexpression of an m6A writer from Solanum lycopersicum, SlHAKAI, could negatively regulate pepino mosaic virus (PepMV) infection, inhibit viral RNA and protein accumulations by affecting viral m6A levels in tomato plants and vice versa. The PepMV-encoded RNA-dependent RNA polymerase (RdRP) directly interacts with SlHAKAI and reduces its protein accumulation. The RdRP-mediated decreased protein accumulation of SlHAKAI is sensitive to the autophagy inhibitor 3-methyladenine and is compromised by knocking down a core autophagy gene. Furthermore, PepMV RdRP could interact with an essential autophagy-related protein, SlBeclin1. RdRP, SlHAKAI, and SlBeclin1 interaction complexes form bright granules in the cytoplasm. Silencing of Beclin1 in Nicotiana benthamiana plants abolishes the RdRP-mediated degradation of SlHAKAI, indicating the requirement of Beclin1 in this process. This study uncovers that the PepMV RdRP exploits the autophagy pathway by interacting with SlBeclin1 to promote the autophagic degradation of the SlHAKAI protein, thereby inhibiting the m6A modification-mediated plant defense responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pepino mosaic virus (PepMV), a member of the genus Potexvirus, was first reported in pepino (Solanum muricatum) in Peru (Jones et al. 1980) and later in tomato crops around the globe (Hanssen and Thomma 2010; Amer and Mahmoud 2020; He et al. 2020). PepMV can infect many Solanaceae species in nature, which causes mild and severe mosaics, bubbling, laminal distortions, and stunting (Hanssen et al. 2008, 2009; Pagan et al. 2006) in leaves or fruit marbling, discoloration, “open fruit” in fruits (Hanssen et al. 2008; Spence et al. 2006). The PepMV genome consists of a single, positive-sense, 6400-nucleotide (nt) RNA strand, encoding a 164-kDa RNA-dependent RNA polymerase (RdRP), three triple gene block (TGB) proteins of 26-, 14-, and 9-kDa (assigned TGBp1, TGBp2, and TGBp3, respectively) and a 25-kDa coat protein (CP) (Gómez et al. 2009). PepMV is highly contagious in tomato plants, as it easily spreads by the standard crop handling procedures through contaminated tools, hands, and clothing and by direct plant-to-plant contact, making it a challenge to tomato production (Spence et al. 2006).
HAKAI, also known as Casitas B-lineage lymphoma-transforming sequence-like protein 1 (CBLL1), was initially identified as a RING finger-type E3 ubiquitin-ligase for the E-cadherin complex in mammals, which bound to the cytoplasmic domain of E-cadherin and mediated its ubiquitination and degradation (Fujita et al. 2002; Pece and Gutkind 2002). Recently, HAKAI as an m6A writer has been confirmed in Arabidopsis and Drosophila (Růžička et al. 2017; Wang et al. 2021; Bawankar et al. 2021). m6A, catalyzed by methyltransferases (writers), removed by demethylases (erasers), and recognized by m6A binding proteins (readers), is the most pivotal internal modification and is widely present in rRNAs, mRNA, tRNAs, miRNA, and long non-coding RNA (Yue et al. 2019). Increasing evidence shows that m6A modification also regulates viral life cycles, and its roles in the arms race of hosts and viruses have been revealed in many reports (Courtney et al. 2017; Ye et al. 2017; Imam et al. 2018; Hao et al. 2019). In the interactions between plants and viruses, AtALKBH9B was first identified as an m6A demethylase in Arabidopsis, which interacted with the coat protein (CP) of Alfalfa mosaic virus (AMV). The m6A methylation of viral RNA limits the infectivity of AMV to the point, where systemic spread into some tissues is nearly entirely blocked if the virus cannot use the host-encoded m6A demethylase to achieve demethylation (Martínez-Péreza et al. 2017). Tobacco mosaic virus (TMV) infection reduced the m6A level in Nicotiana tabacum and promoted ALKBH5-dependent m6A demethylation using a UHPLC-HR-MS/MS method (Li et al. 2018b). A recent report revealed the dynamics of m6A modification during the interaction between rice and rice stripe virus (RSV) or rice black-streaked dwarf virus (RBSDV), which may act as a primary regulatory strategy in gene expression (Zhang et al. 2021). However, whether and how m6A modification regulates virus infection in tomato plants has not been investigated.
Autophagy is a highly conserved intracellular degradation system in eukaryotes for the degradation and recycling of cytoplasmic damaged proteins and cellular organelles (Klionsky and Codogno 2013; Lamb et al. 2013). So far, a series of autophagy-related genes (ATGs) have been identified in plants (Thompson and Vierstra 2005; Yoshimoto et al. 2010; Zhou et al. 2015). ATGs function individually or synergistically during the initiation, elongation, and closure of autophagosome formation (Ding et al. 2018; Soto-Burgos et al. 2018). In plants, autophagy was consolidated as a critical component of plant antiviral immunity, which targets viral proteins to autophagosomes for degradation (Nakahara et al. 2012; Hafren et al. 2017, 2018; Haxim et al. 2017; Li et al. 2018a). However, recent reports have revealed that plant viruses could also manipulate or inhibit autophagy via different strategies to counter plant defense (Li et al. 2017, 2020; Yang et al. 2018, 2022; Zhou et al. 2021). Whether plant viruses could manipulate autophagy to counteract m6A modification-mediated defense responses in the arms race of host-virus is largely obscure.
In this study, we demonstrate that SlHAKAI from Solanum lycopersicum, an m6A writer, is involved in anti-PepMV defense in tomato plants. The RdRP encoded by PepMV could interact with SlHAKAI and promote its protein degradation. Furthermore, we show that the PepMV RdRP exploits the autophagy pathway by directly interacting with SlBeclin1 to mediate the autophagic degradation of SlHAKAI. These results suggest that a viral protein could exploit the autophagy factor to compromise the m6A-mediated antiviral response, which is a novel virulence strategy during the arms race between plants and viruses.
Materials and methods
Plant materials and growth conditions
Seeds of tomato (Solanum lycopersicum) and Nicotiana benthamiana from our lab were cultivated in soil and incubated in an insect-free growth chamber at 25 °C and 40% relative humidity under a 16 h/8 h light/dark photoperiod. The transgenic RFP-H2B line was a gift from Michael M. Goodin (University of Kentucky, USA). The tomato variety used in this study was Micro-Tom (MT), and the SlHAKAI overexpression and SlHAKAI CRISPR/Cas9 mutant plants were generated in an MT background.
Plasmid construction and plant transformation
The Gateway system (Invitrogen) and the enzyme digestion connection method were used to construct recombinant plasmids. The full-length coding sequences of SlHAKAI and its truncated mutants SlHAKAI-N, SlHAKAI-M, and SlHAKAI-C were cloned by RT-PCR from the cDNA derived from Solanum lycopersicum leaves, while PCR cloned the full-length RdRP and its domains Met, Hel, and RdRP2 of PepMV (KY031324) from PepMV infectious clone previously constructed using the specific primer pairs. (Supplementary Table 1). dGDD, the mutant of RdRP2 lacking the GDD motif, was cloned by overlapping PCR with primers listed in Supplementary Table 1. Briefly, the genes mentioned above were amplified by PCR using TransStart® FastPfu DNA Polymerase (TransGen Biotech, China). The resulting DNA fragments were purified (E.Z.N.A. Gel Extraction Kit) and transferred into the entry vector pDONR221 (Invitrogen) by recombination using BP Clonase® (Invitrogen). Insertions in the resulting pDONR clones were verified by sequencing. Then, the linearized fragments using the restriction endonuclease MluI were transferred into modified Gateway-compatible and gateway-compatible vectors to generate corresponding expression vectors. These plasmids, including pGADT7-DEST (AD), pGBKT7-DEST (BD), pEarleygate201-YN, pEarleygate202-YC, pEarleyGate-101 (101, C-terminal YFP), pEarleyGate102 (CFP in the C terminal), pEarleyGate104 (YFP in the N terminal) and pBA-Flag-Myc4 (Myc tag in the N terminal), were constructed in this study. TRV-based recombinant VIGS vectors (pTRV2-NbATG7, pTRV2-NbBeclin1) were described in our previous report (Li et al. 2018a). To obtain the SlHAKAI overexpression vectors, the full-length of SlHAKAI purified above was digested with restriction endonucleases Kpn I/BamH I and ligated to the linearized pCambia1300-GFP (C-terminal GFP) with T4 DNA ligase (TransGen Biotech, China). The SlHAKAI-Cas9-sgRNA construct was obtained as followings: the ideal spacer was referred to the CRISPR-P online website (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). SgRNA sequence is CTTCCGGTAGCCAAGAGCCTTGG. SlHAKAI-spacer1-F/R primers were designed according to the sequence of sgRNAs (Supplementary Table 1). After annealing, the double fragments were directly connected with the BKG01 vector (linearized in advance by Eco31 I) to obtain BKG01-SlHAKAI. After DNA sequencing was confirmed, Agrobacterium-mediated tomato transformation was carried out at BIOGLE Gene Tech Co., Ltd. (Jiangsu, China).
Agroinfiltration and viral inoculation
For viral infection analysis, 3-week-old tomato plants were used for virus inoculation. The infectious clones of PepMV were transformed into A. tumefaciens (strain GV3101) by electroporation. The transformed Agrobacterium cultures were grown individually until approximately OD600 = ~ 2.0. Then, the cultures were collected and re-suspended to OD600 = 1.0 using infiltration buffer (10 mM MgCl2, 100 mM MES (pH 5.7), 2 mM acetosyringone) for 3 h at room temperature. The suspensions were infiltrated into cotyledons of tomatoes using 1-mL syringes. Inoculated plants were photographed with a Canon 400D digital camera at different periods. The PepMV infection assay was repeated at least three times. For transient expression analysis in N. benthamiana leaves, constructs generated were transformed into A. tumefaciens strain EHA105 via electroporation. The wild-type or RFP-H2B transgene N. benthamiana plants in the 4–5 leaf stage were used for Agrobacterium-mediated transient expression with minor modifications of our previous procedures (Li et al. 2018a). Agrobacterium cultures harboring pTRV1 and pTRV2-VIGS (TRV2-GUS, TRV2-NbATG7, TRV2-NbBeclin1) were resuspended for the TRV-VIGS assay in infiltration buffer and mixed at a 1:1 ratio. The mixed A. tumefaciens cultures were infiltrated into leaves of N. benthamiana plants at the 4–5 leaf stage after 3 h incubation at room temperature. As an indicator, N. benthamiana plants infiltrated with TRV-based VIGS vectors targeting phytoene desaturase (PDS). TRV-GUS- or TRV-VIGS-treated plants were agroinfiltrated with PepMV infectious clones when the silenced phenotype of TRV-PDS-treated plants appeared.
Y2H, BiFC, subcellular localization
Y2H was performed according to the Clontech yeast protocol. Briefly, plasmids expressing viral and host proteins were co-transformed into yeast cells (strain Y2H Gold, Clontech catalog number: 630498). Then, selective culture mediums lacking tryptophan and leucine (SD/-Trp-Leu) or tryptophan, leucine, histidine, and adenine (SD/-Trp-Leu-His-Ade) were plated with yeast cells mentioned above to confirm the right transformation or analyze the interactions. BiFC and subcellular localization experiments were performed as previously described (Li et al. 2018a). After 36–72 h infiltration, 1–2 cm2 leaf sections were excised for examining fluorescence in epidermal cells by confocal microscopy (Carl Zeiss 980, German), equipped with a 63 × water-corrected objective in multitrack mode. CFP was excited at 458 nm and captured at 470–500 nm, YFP was excited at 514 nm and captured at 565–585 nm, and RFP was excited at 543 nm and captured at 590–630 nm. In general, at least 20 cells were examined for each experiment. The sequential scanning mode was applied for the co-imaging of different fluorescent proteins. Images were collected and analyzed using ZEN 2 (Carl Zeiss Microscope GmbH2011) imaging software.
RNA extraction and qRT-PCR analysis
The total RNAs of plant leaves were extracted using TRIzol reagent (Invitrogen, USA, Cat. no. 15596-026) according to the manufacturer’s protocols. The extracted RNAs were then reverse-transcribed with PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa) using oligo (dT) primers after removing gDNA. cDNA synthesized from reverse transcription of RNA samples was used to determine the mRNA expression levels of target genes and to quantify PepMV accumulation levels at the date indicated. NbActin or SlActin was used as an internal control for N. benthamiana or tomato. Primers used in this study are provided in Supplementary Table 1.
Western blotting analysis
The plant tissues to be tested were extracted with SDS lysis buffer (100 mM Tris–HCl, pH = 6.8, 10% SDS) for Western blotting analysis as previously described (Li et al. 2018a). Western blotting was performed with primary mouse polyclonal antibodies, followed by goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Bioeasytech, no. YM6704). Anti-PepMV CP polyclonal antibody at 1:10,000 dilution was used for diagnosis of PepMV-infection. Anti-GFP antibody (1:5000; Abcam, no. ab6556) and anti-Myc antibody (1:5000; Abcam, no. ab32) were used to diagnose the target protein. Blotted membranes were washed 3 times (5 min each time) thoroughly with PBST buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4, 0.05% Tween 20) and visualized using chemiluminescence according to the manufacturer’s protocol (ECL; GE Healthcare). Total proteins were stained with Coomassie Brilliant Blue R-250 (CBB) to show equal loading. The experiments in this study were repeated at least three times with similar results.
Chemical treatments
Phosphate-buffered saline buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4) containing 2% dimethyl dulfoxide (DMSO; control) or 2% 5 mM 3-MA (Sigma) for inhibition of autophagy, or 2% (100 µM) MG132 (Sigma-Aldrich) for inhibition of the 26S proteasome, was infiltrated into leaves 8–12 h before leaves were collected.
Extraction of PepMV particles
Nearly 20 g of tomato leaves infected with PepMV were used for viral particle extraction at 15 dpi. The fresh leaves and 40 mL lysis buffer (0.5 mol/L Sodium Phosphate Buffer (pH 7.5), 0.01 mol/L Na-EDTA, 0.1% β-mercaptoethanol) were mixed and grounded with a homogenizing machine for 5 min. The mixture was further filtered with double-layer gauze and centrifuged for 20 min at 6000 r/min. 7% PEG, 2.5% Triton X-100 and 0.1 mol/L NaCl were subsequently added to the supernatant obtained in the last step and stirred at 4 °C for 4 h. Then, the mixture was centrifuged for 15 min at 11,000 r/min, and the precipitation was retained and washed 3 times with 0.02 mol/L Sodium phosphate buffer (PH 7.5). The supernatant obtained in the last step was then added to the 35-mL unique thin-wall centrifugal tube for ultra-speed centrifuge with a 5 mL 30% sucrose pad at the bottom of the tube and subjected to 35,000 r/min centrifugation for 100 min. The precipitation was resuspended with 0.02 mol/L sodium phosphate buffer (PH 7.5), and the resuspension was just the crude extract of PepMV particles.
m6A dot blot assay
First, RNA was made a serial dilution to 400 ng/μL, 200 ng/μL, and 50 ng/μL using RNase-free water (PepMV particles to 1 µg/μL, 0.5 µg/μL, 0.2 µg/μL with 0.02 mol/L sodium phosphate buffer) and denatured at 90 °C for 3 min. Chilling on ice immediately after denaturation, 2 μL mRNA or PepMV particles were dropped on the Hybond-N + membrane optimized for nucleic acid transfer. Then, the membrane was cross-linked in a Stratalinker 2400 UV Crosslinker by UV light twice using the Autocrosslink mode for 1 min, washed with 10 mL wash buffer (1 × PBS, 0.02% Tween-20), and incubated in 10 ml of blocking buffer (1 × PBS, 0.02% Tween-20, 5% non-fat milk) for 1 h at room temperature with gentle shaking. Afterward, the membrane was incubated with the anti-m6A antibodies (1:250 dilution; 2 μg/mL) in 10 mL of antibody dilution buffer (1 × PBS, 0.02% Tween-20, 5% non-fat milk) overnight at 4 °C with gentle shaking. At last, the imaging system visualized the membrane after incubation with the secondary antibodies for 1 h at room temperature. RNA was stained with 0.1% Methylene blue (MB; Solarbio, China, Cat#G1300), while PepMV particles were blotted with PepMV CP antibodies to show equal loading. The experiments in this study were repeated at least three times with similar results.
Results
SlHAKAI is involved in antiviral defense in Solanum lycopersicum
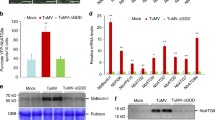
Although MTA, MTB, VIRILIZER, and FIP37 are essential members of the m6A writer complex, knockout of MTA, MTB, VIRILIZER, or FIP37 led to embryo lethal in Arabidopsis (Růžička et al. 2017). In this study, we either failed to get the MTA-, MTB-, VIRILIZER-, or FIP37-knockout tomato plants. Thus, we chose HAKAI as the research object. HAKAI is a conserved component of the methyltransferase complex in eukaryotes (Bawankar et al. 2021). In Arabidopsis, HAKAI is also functionally required for complete m6A mRNA methylation (Růžička et al. 2017). By constructing evolutionary trees and analyzing functional domains of the HAKAI proteins in different plant species, we found that the HAKAIs from several plant species share a high identity in amino acid sequences. All of them have a conserved RING domain (Fig. 1A, B). To investigate whether HAKAI influences PepMV infection in tomato plants, SlHAKAI-GFP transgenic plants were generated and obtained via the Agrobacterium-mediated leaf disc method (Sun et al. 2006). Line 3 (SlHAKAI-GFP-3) and line 8 (SlHAKAI-GFP-8) of the SlHAKAI-overexpression tomato plants showed high levels of SlHAKAI-GFP by Western blot analysis using anti-GFP antibodies (Fig. 1D and Fig. S1B). At the same time, we used CRISPR/Cas9 technology to obtain SlHAKAI-knockout tomato lines (slhakai-1 and slhakai-4). slhakai-1 carried five nucleotide deletions (Fig. 1E), while slhakai-4 carried one nucleotide insertion near the cleavage site (Fig. S1C). SlHAKAI-overexpression and SlHAKAI-knockout tomato plants displayed similar growth, development, and fruit phenotypes with wild-type (WT) tomato plants (Fig. 1C, Fig. S1A, and data not shown), which was consistent with the fact that both two hakai alleles resemble WT in Arabidopsis plants (Růžička et al. 2017). In T2 generation tomato plants, overexpression of SlHAKAI and knockout of SlHAKAI increased and decreased the total m6A levels compared to WT using m6A dot blotting analysis, respectively (Fig. 1F). This result confirmed that SlHAKAI played a role in m6A modification in tomato plants. Then, the SlHAKAI-GFP transgenic and slhakai tomato plants in the T2 generation were inoculated with PepMV infectious clones by agroinfiltration. The inoculated plants were maintained to monitor symptom development. After inoculating these plants with PepMV at 15 days, WT tomato plants showed typical and obvious mosaic symptoms (Fig. 1G and Fig. S1D). SlHAKAI-overexpression plants displayed delayed symptom development and milder mosaic phenotypes than WT plants (Fig. 1G and Fig. S1D). In contrast, significant severe mosaic leaf symptoms appeared on the SlHAKAI-knockout tomato plants (Fig. 1G and Fig. S1D). The newly emerged leaves with mosaic symptoms were then extracted for RNA and protein. As shown in Fig. 1H, I, fewer viral RNA and coat protein (CP) accumulations of PepMV were found in SlHAKAI-overexpression transgenic tomato plants compared to the WT plants. Consistently, more viral RNA and CP accumulated in the SlHAKAI-knockout tomato plants compared to the WT plants (Fig. 1H, I and Fig. S1E). Furthermore, the viral particles from PepMV-infected WT, SlHAKAI-overexpression, and SlHAKAI-knockout tomato plants were extracted and purified for viral genome m6A detection at 15 dpi. Dot blot analysis using anti-m6A antibodies revealed that the viral RNA from SlHAKAI-overexpression and SlHAKAI-knockout tomato plants exhibited more and fewer m6A levels than that from WT plants, respectively (Fig. 1J). These findings indicate that SlHAKAI negatively regulates PepMV infection, probably through m6A modifications of viral RNA.
SlHAKAI negatively regulates tomato resistance to PepMV. A Phylogenetic analysis is shown for amino acid sequences of HAKAI proteins from 8 representative plant species. Sequence alignments and tree construction were conducted in MEGA7 using the Neighbour-Joining method. NbHAKAI-1, 2, HAKAIs from Nicotiana benthamiana; SlHAKAI, HAKAI from Solanum lycopersicum; GmHAKAI-1, 2, HAKAIs from Glycine max; BrHAKAI, HAKAI from Brassica rapa; AtHAKAI, HAKAI from Arabidopsis thaliana; ZmHAKAI, HAKAI from Zea mays L.; OsHAKAI, HAKAI from Oryza sativa L.; TaHAKAI-1, 2, HAKAIs from Triticum aestivum L. B Domain compositions of HAKAI proteins in AtHAKAI, SlHAKAI, OsHAKAI, and NbHAKAI. All HAKAI proteins contain a conserved RING domain. C The phenotypes of SlHAKAI-overexpression (SlHAKAI-GFP-3) transgene and SlHAKAI-knockout (slhakai-1) plants in the T2 generation. WT: wild-type (WT). The indicated plants were photographed 30 days after seeding. White bar represents 5 cm. D Western blot analysis of the SlHAKAI-GFP protein accumulation in SlHAKAI-GFP-overexpression transgenic tomato plants using anti-GFP antibodies. SlHAKAI-GFP-1, 2, 3, 4, 5 represents the 5 individual transgene lines. Coomassie Brilliant Blue R-250 (CBB)-stained Rubisco large subunit was set as a loading control. E DNA sequencing and sequence alignment were conducted to confirm the mutation in the SlHAKAI-knockout (slhakai-1) transgene plants. This nucleotide deletion caused a frameshift mutation of SlHAKAI. F Dot blotting analysis of the overall levels of m6A modification in WT, SlHAKAI-GFP-3, and slhakai-1 tomato plants using anti-m6A antibodies. Methylene blue (MB) staining of total RNA served as an equal loading. G The pepino mosaic virus (PepMV) caused symptoms in WT, SlHAKAI-GFP-3, and slhakai-1 tomato plants. The PepMV-inoculated tomato plants were photographed at 15 dpi. The white bar represents 2 cm. H Relative viral accumulations of PepMV in the plants shown in (G) were detected by qRT-PCR at 15 dpi. SlActin was used as the internal reference gene to normalize the relative expression, and the value in WT tomato plants was set to 1. Values represent the mean ± SD from 3 independent biological samples. Student’s t-test was used to analyze each data group, and double asterisks indicate significant statistical differences (**P < 0.01) between the two treatments. I The accumulation levels of PepMV protein in (G)-indicated plants were determined by Western blotting using anti-PepMV CP antibodies at 15 dpi. CBB-stained Rubisco large subunit was a loading control. J Dot blotting was used to detect m6A modification levels with PepMV particles extracted from PepMV-infected WT, SlHAKAI-GFP-3, and slhakai-1 plants indicated in (G) using anti-m6A antibodies. Blotting with antibodies of PepMV CP indicated the loading amount of PepMV particles
PepMV RdRP interacts with SlHAKAI
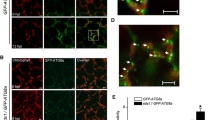
SlHAKAI played an anti-viral role during PepMV infection, so we wondered whether viral proteins could inhibit or manipulate SlHAKAI to counter-defend SlHAKAI-mediated anti-viral responses. Thus, possible interactions between SlHAKAI and the 5 PepMV-encoded viral proteins were screened by yeast two-hybrid (Y2H) assays. Positive protein–protein interaction was found between SlHAKAI and PepMV RdRP, and no interaction was observed between SlHAKAI with any other PepMV proteins (Fig. 2A and Fig. S2A). The RdRP-SlHAKAI interaction was further examined by bimolecular fluorescence complementation (BiFC) in RFP-H2B (a nuclear marker) transgenic Nicotiana benthamiana leaves. The BiFC assays of the interaction between YN-RdRP and YC-SlHAKAI, or YN-SlHAKAI and YC-RdRP was observed by confocal microscopy at 48 hpi, which formed bright granules in the cytoplasm (Fig. 2B, lines 1, 2). No interaction was observed between SlHAKAI and TGBp3 (PepMV TGBp3, as a negative control) (Fig. 2B, line 3). The BiFC assays of the interaction between YN-SlHAKAI and YC-RdRP upon PepMV infection were also conducted. We found that PepMV infection did not affect the formation of the SlHAKAI-RdRP interaction granules in the cytoplasm (Fig. S3A). SlHAKAI-CFP formed specific punctate-structure dots in the cytoplasm and nucleus, whereas RdRP was uniformly distributed in the cytoplasm and nucleus (Fig. 2C, lines 1, 2). When SlHAKAI-CFP and YFP-RdRP were co-expressed, the localization of YFP-RdRP was redistributed to the SlHAKAI-CFP-labeled punctuate structures in the cytoplasm and nucleus at 48 hpi (Fig. 2C, line 3). In addition, PepMV infection did not influence the localization of SlHAKAI and RdRP either (Fig. S2C). These results suggested that PepMV RdRP could interact with SlHAKAI to form an aggregated interaction complex in the cytoplasm and nucleus in planta.
SlHAKAI interacts with PepMV RdRP. A Y2H assays of the interaction between SlHAKAI and RdRP. Yeast cells co-transformed with AD-T7-T + BD-T7-53 serve as a positive control; yeast cells co-transformed with AD-SlHAKAI and the empty BD, with the empty AD and BD-RdRP, or with AD-T + BD-Lam are negative controls. B Confirmation of the SlHAKAI-RdRP interaction by BiFC assay. BiFC assays between SlHAKAI and RdRP in the leaves of RFP-H2B (red) transgenic N. benthamiana. Confocal imaging was performed at 48 hpi. SlHAKAI and RdRP were fused to the N (YN) and C-terminal (YC) fragments of yellow fluorescent protein (YFP). The yellow fluorescence indicated the SlHAKAI-RdRP. Bars, 10 μm. C Co-localization of SlHAKAI-CFP and YFP-RdRP in the leaf cells of RFP-H2B transgenic N. benthamiana by confocal microscopy at 48 hpi. Arrows indicate the overlapping fluorescence of SlHAKAI-CFP and YFP-RdRP. Bars, 10 μm. D Schematic representation of full-length and truncated proteins of SlHAKAI and RdRP. The conserved domains in SlHAKAI and RdRP are indicated. RING: RING Ubox domain; Met: methyltransferase domain; Hel: helicase domain; RdRP2: RdRP2 domain. E Mapping the interaction domain between SlHAKAI and RdRP by Y2H assays. Y2H Gold yeast cells co-transformed with the indicated plasmids were subjected to tenfold serial dilutions and plated on synthetic dextrose (SD)/-Trp, -Leu, -His, -Ade or SD/-Trp, -Leu medium to screen for possible interactions at 3 days after transformation (A and E). F BiFC assays of the domains of SlHAKAI and RdRP. Confocal images were taken at 48 h post infiltration (hpi). The nuclei of N. benthamiana leaf epidermal cells are marked by RFP-H2B (red). These experiments were repeated three times independently. At least 20 cells per sample were observed, and representative results were displayed. Bars, 10 μm (B, C, and F)
We next mapped the protein domains required for the interaction between SlHAKAI and RdRP. SlHAKAI contains a conserved N-terminal RING domain, while RdRP contains three conserved domains, including Met (the viral methyltransferase domain), Hel (the viral RNA helicase domain), and RdRP2 (the viral RdRP2 domain) (Fig. 2D). Then, SlHAKAI and RdRP were divided into three fragments based on their domain compositions (SlHAKAI- N, M, C, and RdRP- Met, Hel, RdRP2). SlHAKAI-N includes a RING domain, and no specific motif exists in SlHAKAI-M or SlHAKAI-C domain (Fig. 2D). Y2H revealed that the N-terminal RING domain of SlHAKAI interacted with the RdRP2 domain of RdRP (Fig. 2E and Fig. S3A). SlHAKAI-N and SlHAKAI displayed similar subcellular localization patterns forming bright cytoplasm spots (Fig. S3B). RdRP and RdRP2 fused YFP both localized in the cytoplasm and nucleus (Fig. S3C). Consistently, PepMV infection did not influence the sublocalization patterns of the domains of SlHAKAI and RdRP (Fig. S3B, C). Since the highly conserved GDD motif required for the RdRP activity is located in the domain of RdRP2 of RdRP (Li et al. 2018a), we tested the importance of this motif to the SlHAKAI-RdRP interaction. As expected, the deletion of GDD (dGDD) abolished the interaction between SlHAKAI and RdRP (Fig. 2E). This observation was consistent with results from BiFC assays in planta (Fig. 2F, line 4).
PepMV RdRP promotes the SlHAKAI degradation via autophagy
To investigate the interaction function between SlHAKAI and RdRP, SlHAKAI-YFP was co-expressed with Myc-GUS (mock) or Myc-RdRP2 in RFP-H2B transgenic N. benthamiana plants. At 48 hpi, the intensity of fluorescence and the size and quantity of granules of SlHAKAI were significantly reduced when co-expressed with Myc-RdRP2, compared with the mock (Fig. 3A). Samples above were then collected to extract RNA and protein for qRT-PCR and immunoblotting analysis, respectively. As shown in Fig. 3B, the protein accumulation of SlHAKAI was significantly decreased when co-expressed with YFP-RdRP2. No significant changes in SlHAKAI mRNA were observed when expressed with Myc-GUS or Myc-RdRP2 (Fig. 3C). Furthermore, a similar reduction was also observed when YFP-RdRP2 was co-expressed with Myc-SlHAKAI (Fig. 3D, E).
RdRP promotes the degradation of SlHAKAI. A Confocal micrographs showing RFP-H2B transgene N. benthamiana leaf cells co-infiltrated with Agrobacterium carrying SlHAKAI-YFP and Myc-GUS or Myc-RdRP2 at 48 hpi. Bars, 10 μm. B Immunoblotting analysis of total protein extracted from leaves indicated in (A) at 48 hpi with anti-GFP or anti-Myc antibodies. C Relative expression levels of SlHAKAI in (A)-indicated leaves at 48 hpi. D Immunoblotting analysis of total protein isolated from leaves co-infiltrated with Agrobacterium carrying Myc-SlHAKAI and YFP or YFP-RdRP2 at 48 hpi with anti-Myc or anti-GFP antibodies. CBB-staining of Rubisco large subunit was set as a loading control (B and D). E Relative expression levels of SlHAKAI in (D)-indicated leaves at 48 hpi. NbActin was used as the internal reference gene to normalize the relative expression, and the value in Myc-GUS and YFP-RdRP2 (C) or YFP and Myc-SlHAKAI (E) co-expressed N. benthamiana leaves was set to 1. Values represent the mean ± SD from 3 independent biological samples. Student’s t-test was used to analyze each data group, and ns represents no significance between the two treatments (C, E)
The most studied proteasome is the primary proteolytic machinery that regulates cellular protein homeostasis (proteostasis) by selectively degrading ubiquitinated proteins (Finley 2009). Given this, drug treatment with MG132, a well-known ubiquitin-26S proteasome system (UPS) inhibitor, was performed to test whether UPS was responsible for the RdRP2-mediated degradation of SlHAKAI. As shown in Fig. 4A, there was no significant difference between DMSO-treated and MG132-treated samples, which indicated that the RdRP2-mediated degradation of SlHAKAI might be UPS-independent. Next, we used an autophagy inhibitor, 3-methyladenine (3-MA), to treat the samples co-expressed with RdRP2 and SlHAKAI. Western blotting analysis revealed that the RdRP2-mediated HAKAI protein degradation was markedly inhibited by treatment with 3-MA (Fig. 4C). No obvious change in SlHAKAI mRNA was observed between DMSO- and 3-MA-treated Myc-SlHAKAI and YFP-RdRP2 co-expressing samples (Fig. 4D). Then, a tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system was employed to silence an essential autophagy gene, NbATG7, to confirm whether autophagy is involved in the RdRP2-mediated degradation of SlHAKAI (Fig. S4A). As shown in Fig. 4E, F, the knockdown of NbATG7 remarkably inhibited the degradation of Myc-SlHAKAI. Besides, we also analyzed the expression levels of SlHAKAI upon PepMV infection when NbATG7 was knocked down. As shown in Fig. 4G, H, PepMV infection reduced the fluorescence intensity and decreased the protein accumulation of SlHAKAI-YFP in TRV-GUS plants. However, the knockdown of NbATG7 (TRV-NbATG7) remarkably inhibited the degradation of SlHAKAI-YFP upon virus infection. Taken together, PepMV infection promoted the autophagic degradation of SlHAKAI, which RdRP was responsible for this through interacting with SlHAKAI.
RdRP mediates the autophagic degradation of SlHAKAI. A MG132 (an inhibitor of the ubiquitin-26S proteasome system) does not affect the RdRP-mediated degradation of Myc-SlHAKAI. Total protein was isolated from plant leaves co-agroinfiltrated with Agrobacterium harboring Myc-SlHAKAI and YFP or YFP-RdRP2, followed by DMSO or MG132 treatment at 48 hpi. B Relative expression levels of SlHAKAI in (A)-indicated leaves at 48 hpi. C Effect of an autophagy inhibitor, 3-MA, on the RdRP-mediated degradation of Myc-SlHAKAI. D Relative expression levels of SlHAKAI in (C)-indicated leaves at 48 hpi. E Effect of silencing of NbATG7 on the RdRP-mediated degradation of Myc-SlHAKAI. Plants preinoculated with TRV-GUS or TRV-NbATG7 at 10 dpi were co-agroinfiltrated with Agrobacterium harboring Myc-SlHAKAI and YFP or YFP-RdRP2 in the upper leaves. Total protein was extracted from infiltrated leaves at 48 hpi. F Relative expression levels of SlHAKAI in (E)-indicated leaves at 48 hpi (B, D, F). G Subcellular localization of SlHAKAI-YFP in the leaf cells of NbATG7-silenced or non-silenced RFP-H2B transgenic N. benthamiana plants with or without PepMV infection by confocal microscopy at 48 hpi. Bars, 10 μm. H Immunoblotting of SlHAKAI-YFP in (G)-indicated leaves at 48 hpi. Immunoblotting was performed using anti-GFP or anti-Myc antibodies. All immunoblotting assays in this figure were repeated at least three times, and one representative blot was shown. CBB-staining of Rubisco large subunit is a loading control (A, C, E, H). I Relative expression levels of SlHAKAI in (E)-indicated leaves at 48 hpi. NbActin was used as an internal control, Student’s t test was used to analyze each data group, and ns represents no significance between the two treatments
The PepMV RdRP-mediated autophagic degradation of SlHAKAI requires SlBeclin1
Our previous report has shown that NbBeclin1 interacted with the RdRPs of several RNA viruses and promoted their degradation (Li et al. 2018a). So, we hypothesized SlBeclin1 might play a role in RdRP-mediated autophagic degradation of SlHAKAI. Y2H assay was conducted to verify whether SlBeclin1 interacted with RdRP. As shown in Fig. 5A, a strong interaction was observed in the selective yeast medium. This interaction was further confirmed by BiFC assay in RFP-H2B transgenic N. benthamiana plants, which revealed that this interaction occurs in the cytoplasm (Fig. 5B). As a negative control, no YFP fluorescence was observed when the movement protein TGBp3 from PepMV was co-expressed with SlBeclin1 or RdRP (Fig. 5B). SlBeclin1 was present in the cytoplasm with specific punctate subcellular localization, while RdRP localized to the cytoplasm and nucleus at 48 hpi when expressed alone (Fig. 5C). However, when YFP-RdRP was co-expressed with SlBecline1-CFP, the localization of YFP-RdRP was re-directed to the SlBeclin1-CFP-labeled punctuate structures in the cytoplasm at 48 hpi (Fig. 5C). Of note, the SlHAKAI-RdRP interaction complex was co-localized with SlBeclin1, forming bright granules in the cytoplasm (Fig. 5D). Since Beclin1 is required for autophagy in mammalian and plant cells (Liang et al. 1999; Fujiki et al. 2007), we thus speculate that Beclin1 may direct the SlHAKAI-RdRP complex into autophagosomes for degradation by interacting with RdRP during viral infection. We then silenced NbBeclin1 using a VIGS vector to determine whether RDRP2-mediated SlHAKAI degradation was affected by the expression of NbBeclin1 (Fig. S4B). As shown in Fig. 5E, F, the RdRP2-mediated degradation of Myc-SlHAKAI was remarkably inhibited in the NbBeclin1-silenced cells.
Beclin1 is required for the RdRP-mediated autophagic degradation of SlHAKAI. A Y2H assays of the interaction between SlBeclin1 and RdRP. Y2H Gold yeast cells co-transformed with the indicated plasmids were subjected to tenfold serial dilutions and plated on a selective medium to screen for positive interactions 3 days after transformation. The yeast cells co-transformed with AD + BD-RdRP, AD-SlBelin1 + BD, or AD-T + BD-Lam were served as negative controls, and AD-T + BD-53 used as a positive control. B Confirmation of SlBeclin1-RdRP interaction by BiFC assay. Bars, 10 µm. C By confocal microscopy, the co-localization of SlBeclin1-CFP and YFP-RdRP in the leaf cells of RFP-H2B transgenic N. benthamiana. Arrow indicates the overlapping fluorescence, which was produced from SlBeclin1-CFP and YFP-RdRP. Bars, 10 µm. D Co-localization of the SlHAKAI-RdRP interaction complex with SlBeclin1. Bars, 10 μm. Confocal images were taken at 48 hpi. The nuclei of N. benthamiana leaf epidermal cells are marked by RFP-H2B (red). These experiments were repeated three times independently. At least 20 cells per sample were observed, and representative results were displayed (B–D). E Effect of silencing of NbBeclin1 on the RdRP-mediated degradation of Myc-SlHAKAI. Plants pre-inoculated with TRV-GUS or TRV-NbBeclin1 at 10 dpi were then co-agroinfiltrated with Agrobacterium harboring Myc-SlHAKAI and YFP or YFP-RdRP2. Total protein was extracted from infiltrated leaves at 48 hpi. Immunoblotting was performed using anti-GFP or anti-Myc antibodies. All immunoblotting assays in this figure were repeated at least three times, and one representative blot was shown. CBB staining of Rubisco large subunit serves as a loading control. F Relative expression levels of SlHAKAI in (E)-indicated leaves at 48 hpi. NbActin was used as an internal control, Student’s t-test was used to analyze each data group, and ns represents no significance between the two treatments
Discussion
Plants’ successful survival depends on their ability to exploit numerous defense mechanisms against invading pathogens or hostile environments. Here, we demonstrate that SlHAKAI is involved in anti-PepMV defense via m6A modification of viral RNA in tomato plants. m6A, the most pivotal internal modification in RNAs, is involved in various biological processes in eukaryotes (Yue et al. 2019). Crosstalk between mRNA m6A modification and autophagy has been reported in mammals (Jin et al. 2018; Wang et al. 2020; Chen et al. 2021). In autophagy, a critical initial event is the formation of the autophagosome. This unique double-membrane organelle engulfs the cytosolic cargo destined for degradation, which is mediated by the serine/threonine protein kinase ULK1 (unc-51-like kinase 1) (Zachari and Ganley 2017). ULK1 mRNA was revealed to undergo m6A modification in the 3′-UTR, and the m6A-marked ULK1 transcripts can further be targeted for degradation by YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). The m6A sites on the transcripts were further determined to be the direct substrates of FTO (fat mass and obesity-associated protein), which removes the m6A mRNA modification of ULK1 transcripts, thus promoting autophagy and prolonging the half-life of ULK1 transcripts (Jin et al. 2018). A further study identified that Atg5 and Atg7 transcripts were the targets of YTHDF2, resulting in mRNA degradation and reduction of protein expression, thereby alleviating autophagy (Wang et al. 2020). However, whether autophagy could regulate the mRNA m6A modification in feedback and affect the m6A-related proteins is not clear. In this study, we demonstrated that PepMV RdRP exploits the autophagy pathway by directly interacting with SlBeclin1 to promote the autophagic degradation of the SlHAKAI protein, which was involved in the m6A modification of viral RNA. Thus, this study provides a novel insight into the interplay between autophagy and m6A methylation.
Autophagy, as a critical component of plant antiviral innate and adaptive immunity, has been consolidated by numerous studies recently (Nakahara et al. 2012; Hafren et al. 2017, 2018; Haxim et al. 2017; Li et al. 2018a). However, some plant viruses can repress or even manipulate autophagy to counter plant defense. For instance, disruption of autophagy by silencing ATG genes ATG7 or ATG8f or treatment with the autophagy inhibitor 3-MA reduces bamboo mosaic virus (BaMV) accumulation, suggesting that autophagy could play a pro-viral role during BaMV infection (Huang et al. 2019). Rgs-CaM, an endogenous RNA silencing suppressor, was reported to promote geminivirus infection by interacting with the suppressor of gene silencing 3 (SGS3) of RNA silencing to mediate its autophagic degradation in N. benthamiana (Li et al. 2017). In another study, the γb protein encoded by barley stripe mosaic virus was revealed to interfere with the interaction of ATG7 and ATG8 in a competitive manner (Yang et al. 2018). Combined with our previous finding (Li et al. 2018a) and this study, we revealed that the PepMV RdRP could become a target of autophagy. However, it could also exploit the autophagy pathway to inhibit m6A modification-mediated anti-viral defense by promoting the degradation of SlHAKAI. Therefore, this finding expanded our understanding of the role of autophagy in the mRNA m6A modification pathway, indicating its perplexing roles in the context of virus infection.
In summary, our study demonstrates that PepMV RdRP exploits the autophagy pathway by directly interacting with SlBeclin1 to promote the autophagic degradation of the SlHAKAI protein, which is involved in m6A modification. Our study highlights the functional importance of the autophagy machinery in regulating m6A modification during viral infection. These findings provide insights into the crosstalk among autophagy, m6A modification, and viral counter-defense.
Data availability
The data sets generated and analyzed during the current study are available from the corresponding author upon request.
References
Amer MA, Mahmoud SY (2020) First report of Tomato brown rugose fruit virus on tomato in Egypt. New Disease Reports 41:24
Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, Heidelberger JB, Richter FM, Nallasivan MP, Morin V, Kreim N, Beli P, Helm M, Jinek M, Soller M, Roignant JY (2021) Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat Commun 12(1):3778
Chen X, Wang J, Tahir M, Zhang F, Ran Y, Liu Z, Wang J (2021) Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases. Cell Biosci 11(1):147
Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR (2017) Epitranscriptomic enhancement of Influenza A Virus gene expression and replication. Cell Host Microbe 22(3):377-386.e5
Ding X, Zhang X, Otegui MS (2018) Plant autophagy: new flavors on the menu. Curr Opin Plant Biol 46:113–121
Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78:477–513
Fujiki F, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 143(3):1132–1139
Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W (2002) Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol 4(3):222–231
Gómez P, Sempere RN, Elena SF, Aranda MA (2009) Mixed infections of pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J Virol 83(23):12378–12387
Hafren A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci USA 114(10):E2026–E2035
Hafren A, Ustun S, Hochmuth A, Svenning S, Johansen T, Hofius D (2018) Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HC-pro. Plant Physiol 176(1):649–662
Hanssen IM, Thomma BP (2010) Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol Plant Pathol 11(2):179–189
Hanssen IM, Paeleman A, Wittemans L, Goen K, Lievens B, Bragard C, Vanachter A, Thomma B (2008) Genetic characterization of Pepino mosaic virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol 121:131–146
Hanssen IM, Paeleman A, Vandewoestijne E, Bergen LV, Bragard C, Lievens B, Vanachter ACRC, Thomma BPHJ (2009) Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol 58:450–460
Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, Wang H, Zhang B, Qiu J, Deng F, Guan WX (2019) N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res 47(1):362–374
Haxim Y, Ismayil A, Jia Q, Wang Y, Zheng X, Chen T, Qian L, Liu N, Wang Y, Han S, Cheng J (2017) Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 6:e23897
He W, Wu J, Ren Y, Zhou X, Zhang S, Qian Y, Li F, Wu J (2020) Highly sensitive serological approaches for Pepino mosaic virus detection. J Zhejiang Univ-SC B 21(10):811–822
Huang Y, Huang Y, Hsiao Y, Li S, Hsu Y, Tsai CH (2019) Autophagy is involved in assisting the replication of Bamboo mosaic virus in Nicotiana benthamiana. J Exp Bot 70:4657–4670
Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, Kim SJ, Mason CE, Horner SM, Siddiqui A (2018) N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci USA 115(35):8829–8834
Jin S, Zhang X, Miao Y, Liang P, Zhu K, She Y, Wu Y, Liu D, Huang J, Ren J, Cui J (2018) m6A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res 28(9):955–957
Jones RAC, Koenig R, Lesemann DE (1980) Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum). Ann Appl Biol 94:61–68
Klionsky DJ, Codogno P (2013) The mechanism and physiological function of macroautophagy. J Innate Immun 5(5):427–433
Lamb CA, Yoshimori T, Tooze SA (2013) The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 14(12):759–774
Li F, Zhao N, Li Z, Xu X, Wang Y, Yang X, Liu S, Wang AM, Zhou X (2017) A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog 13:e1006213
Li F, Zhang C, Li Y, Wu G, Hou X, Zhou X, Wang AM (2018a) Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat Commun 9(1):1268
Li Z, Shi J, Yu L, Zhao X, Ran L, Hu D, Song B (2018b) N6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. J Virol 15(1):87
Li F, Zhang C, Tang Z, Zhang L, Dai Z, Lyu S, Li Y, Hou X, Bernards M, Wang AM (2020) A plant RNA virus activates selective autophagy in a UPR-dependent manner to promote virus infection. New Phytol 228(2):622–639
Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V (2017) Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci USA 114:10755–10760
Nakahara KS, Masuta C, Yamada S, Shimura H, Kashihara Y, Wada TS, Meguro A, Goto K, Tadamura K, Sueda K, Sekiguchi T, Shao J, Itchoda N, Matsumura T, Lgarashi M, Ito K, Carthew RW, Uyeda I (2012) Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci USA 109(25):10113–10118
Pagan I, Cordoba-Selles M, Martinez-Priego L, Fraile A, Malpica J, Jorda C, Garcia-Arenal F (2006) Genetic structure of the population of Pepino mosaic virus infecting tomato crops in Spain. Phytopathology 96:274–279
Pece S, Gutkind JS (2002) E-cadherin and Hakai: signalling, remodeling or destruction? Nat Cell Biol 4(4):E72–E74
Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, De Jaeger G, Mongan NP, Hejátko J, Helariutta Y, Fray RG (2017) Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol 215:157–172
Soto-Burgos J, Zhuang X, Jiang L, Bassham DC (2018) Dynamics of autophagosome formation. Plant Physiol 176:219–229
Spence NJ, Basham J, Mumford RA, Hayman G, Edmondson R, Jones DR (2006) Effect of Pepino mosaic virus on the yield and quality of glasshouse-grown tomatoes in the UK. Plant Pathol 55:595–606
Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for micro-tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47(3):426–431
Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8:165–173
Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q, Shi H, Wang F, Wang Y (2020) m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 16(7):1221–1235
Wang Y, Zhang L, Ren H, Ma L, Guo J, Mao D, Lu Z, Lu L, Yan D (2021) Role of Hakai in m6A modification pathway in Drosophila. Nat Commun 12(1):2159
Yang M, Zhang Y, Xie X, Yue N, Li J, Wang XB, Han C, Yu J, Liu Y, Li D (2018) Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7–ATG8 interaction. Plant Cell 30:1582–1595
Yang M, Wang Y, Li D, Liu Y (2022) Plant virus infection disrupts vacuolar acidification and autophagic degradation for the effective infection. Autophagy 18(3):705–706
Ye F, Chen ER, Nilsen TW (2017) Kaposi’s Sarcoma-associated Herpesvirus utilizes and manipulates RNA N6-Adenosine methylation to promote lytic replication. J Virol 91(16):e00466-e517
Yoshimoto K, Takano Y, Sakai Y (2010) Autophagy in plants and phytopathogens. FEBS Lett 584:1350–1358
Yue H, Nie X, Yan Z, Weining S (2019) N6-methyladenosine regulatory machinery in plants: composition, function and evolution. Plant Biotechnol J 17(7):1194–1208
Zachari M, Ganley IG (2017) The mammalian ULK1 complex and autophagy initiation. Essays Biochem 61(6):585–596
Zhang K, Zhuan X, Dong Z, Xu K, Chen X, Liu F, He Z (2021) The dynamics of N6-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol 22(1):189
Zhou X, Zhao P, Wang W, Zou J, Cheng T, Peng X, Sun M (2015) A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res 22(4):245–257
Zhou T, Zhang M, Gong P, Li F, Zhou X (2021) Selective autophagic receptor NbNBR1 prevents NbRFP1-mediated UPS-dependent degradation of βC1 to promote geminivirus infection. PLoS Pathog 17(9):e1009956
Acknowledgements
This work was funded by the National Key Research and Development Program of China (2021YFD1400400) to FL and the National Natural Science Foundation of China (31930089 and 31972244) to XZ and FL.
Author information
Authors and Affiliations
Contributions
FL and XZ designed the project; HH, LG, and ZL carried out experiments; all authors analyzed and discussed the data; HH, FL, and XZ wrote the manuscript; all the authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical requirements
This article does not contain any studies with human or animal subjects.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, H., Ge, L., Li, Z. et al. Pepino mosaic virus antagonizes plant m6A modification by promoting the autophagic degradation of the m6A writer HAKAI. aBIOTECH 4, 83–96 (2023). https://doi.org/10.1007/s42994-023-00097-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42994-023-00097-6