Abstract
Grayscale statistical attributes analysed for 513 extract images taken from pulmonary computed tomography (CT) scan slices of 57 individuals (49 confirmed COVID-19 positive; eight confirmed COVID-19 negative) are able to accurately predict a visual score (VS from 0 to 4) used by a clinician to assess the severity of lung abnormalities in the patients. Some of these attributes can be used graphically to distinguish useful but overlapping distributions for the VS classes. Using machine and deep learning (ML/DL) algorithms with twelve grayscale image attributes as inputs enables the VS classes to be accurately distinguished. A convolutional neural network achieves this with better than 96% accuracy (only 18 images misclassified out of 513) on a supervised learning basis. Analysis of confusion matrices enables the VS prediction performance of ML/DL algorithms to be explored in detail. Those matrices demonstrate that the best performing ML/DL algorithms successfully distinguish between VS classes 0 and 1, which clinicians cannot readily do with the naked eye. Just five image grayscale attributes can also be used to generate an algorithmically defined scoring system (AS) that can also graphically distinguish the degree of pulmonary impacts in the dataset evaluated. The AS classification illustrated involves less overlap between its classes than the VS system and could be exploited as an automated expert system. The best-performing ML/DL models are able to predict the AS classes with better than 99% accuracy using twelve grayscale attributes as inputs. The decision tree and random forest algorithms accomplish that distinction with just one classification error in the 513 images tested.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
COVID-19 has wreaked havoc around the globe with huge loss of life, lock downs and consequential economic damage. Analytical methods that help to determine and rapidly classify how severe the pulmonary damage is in patients suffering from the disease is essential for understanding their treatment requirements and accelerating its implementation. Computed tomography (CT) scan-slice images are now an established modality of pulmonary examination. CT data plays an important role in determining the severity of lung conditions associated with several serious diseases, such as cancer, different types of pneumonia and, since 2020, COVID-19 [1, 2]. Radiological investigation techniques (X-ray and CT) offer a means of diagnosing COVID-19 that can accurately complement the results of the virus' nucleic acid by real-time reverse transcription polymerase chain reaction (rRT-PCR), that is the standard laboratory test for the disease [3, 4]. CT scan slices examining a patient’s thorax have become the preferred image modality for detecting and verifying COVID-19 [5].
Research emphasis to date has been mainly associated with the use of CT slices for COVID-19 diagnosis and/or for prognosis to qualitatively monitor the progress of the disease during patient treatment, rather than attempting to quantify the degree of severity of its pulmonary impacts [6]. Nevertheless, the ability of CT scan data to identify the degree of COVID-19 impacts has been exploited by some researchers [7]. Challenges faced in doing this automatically from CT image analysis, with the aid of machine learning and/or deep learning (ML/DL) methods, have been identified [8, 9].
COVID-19 is characterized by some distinctive features in pulmonary CT-scan slices. These include, depending on severity, granular opaqueness, chaotically arranged lineation patterns, agglomerated masses, alveoli becoming opaque, inverse halos or atoll shapes, and thickened polygonal forms with poorly defined linear opacities [10]. However, radiological characterization of various lung pathologies [11] suggests that pre-existing pulmonary abnormalities are likely to complicate COVID-19 diagnosis using CT scans. Attempts to link the deep features discernible in CT scan images have proved worthwhile in severity classification of pulmonary impact in those afflicted with COVID-19 [12]. ML/DL algorithms can be configured to detect these characteristic COVID-19 pulmonary features with meaningful accuracy [13,14,15]. DL techniques that customize convolutional neural networks (CNN) adding automated feature-extraction algorithms are being exploited to good effect [16, 17]. To automatically extract deep features from pulmonary CT scan images it is necessary to effectively apply image segmentation algorithms [14, 18, 19].
Many researchers are now striving to customise pulmonary CT scan image analysis as part of the battle to improve COVID-19 patient diagnosis and prognosis. Some are adapting CNN models linked with reliable feature-extraction algorithms [8]. Such methods can be effective in determining the degree of pulmonary impacts, but often fail to consider the relationships among the grayscale-image properties that are responsible for making the distinctive characteristics visible in CT images. This study, for the first time, specifically addresses these underlying image attributes to transparently determine their influence on CT images associated with different degrees of pulmonary impacts in COVID-19 patients compared to individuals unaffected by the disease. Such information is used to verify the visual assessments of CT scan slices by clinicians. It is also exploited to derive accurate algorithmic classification systems that, with refinements, could form the basis of automated expert systems. Such algorithmic systems offer the potential to automatically classify the degree of severity of pulmonary impacts in COVID-19 patients, and those suffering from other pulmonary conditions.
Analysis of grayscale statistical attributes of CT-image-slice extracts from the pulmonary parenchyma (i.e. the alveolar tissue involved in respiration) forms the focus of this study. Such images can be rapidly extracted and processed to provide their statistical attribute values. Statistical and graphical techniques applied to the grayscale attributes of the image extracts reveal overlapping distributions, some of which are strongly correlated with the severity of lung abnormalities. Supervised learning using a suite of ML/DL algorithms is then applied to predict with high accuracy the degrees of pulmonary impacts associated with CT- image slices using multiple grayscale image attributes as input variables.
Methods
Acquisition of Scan Slices by Computed Tomography
For the research presented, thoracic CT-image slices were collected for multiple individuals, many afflicted with COVID-19, and some unaffected by COVID-19, but all being treated at a hospital in Shiraz (Iran). A Philips Ingenuity CT scanning machine at the Namazi medical centre was used to obtain multiple CT scan slices of 0.625 mm thickness from each patient. Multi-slice CT machines provide rapid and comprehensive imaging and are now widely used [20].
Non-contrasted CT scan images were compiled for this study. Such non-contrasted CT images are now routinely used to provide rapid ongoing assessments of several serious pulmonary disorders including coronavirus. The CT image information usefully complements the results of rRT-PCR tests in the definitive diagnosis of COVID-19. CT image scan slices were selected from forty-nine COVID-positive patients exhibiting a wide range of lung abnormalities, together with CT images from eight COVID-negative patients. The eight COVID-negative patients comprised 4 women (aged 18–68 years) and 4 men (aged 24–71 years). The Covid-positive patients comprised 23 women (aged 22–74 years) and 26 men (aged 32–86 years). Thus, the patients considered cover a balanced distribution of gender and age.
Analysis Conducted on CT Scan Slices
Visual Classification
The CT-image slices collected for every patient included in the dataset were visually scrutinized by a clinician. On the basis of that inspection specific images could be assigned five distinct classifications (identified by the numbers zero to four), in which:
Class 0 consists of patients testing negative for COVID-19 and with no visual signs of other pulmonary abnormalities;
Classes1 to 4 consist of patients testing positive for COVID-19 with CT scan images showing varying degrees of lung abnormalities;
Class 1 patient scans display no or trace visual signs of lung abnormalities;
Class 2 patient scans display clear but minor visual signs of lung abnormalities;
Class 3 patient scans display substantial visual signs of lung abnormalities;
Class 4 patient scans display severe visual signs of lung abnormalities.
Such visual scoring (VS) is then used as the prediction goal for image-extract statistical analysis. Figure 1 displays example CT scan slices of the VS classes 0, 2, 3 and 4 (VS class 1 is not shown because it appears on visual inspection the same as VS class 0). Figure 2 displays rectangular extract images from these example CT scans used for statistical analysis. It takes a few seconds to capture each image extract, and a few more seconds to determine and record a range of statistical attributes from each image.
Example CT extract images (enlarged) for patients in the dataset assigned to: A VS class 0 (COVID-negative) with no or trace visual lung abnormalities; B VS class 2 with minor but distinct abnormalities; C VS class 3 with substantial visual abnormalities; and, 4) VS class 4 with severe visual abnormalities. These particular extracts are taken from the CT scan slices shown in Fig. 1A–D, respectively
Extract Image Selection and Analysis
From the inspection of the CT-scan-slice collection for each individual studied, several slices were selected for detailed analysis to best represent their pulmonary condition. One quadrilateral extract was sampled from each lung in each CT slice selected. This accumulated three hundred and ninety two image extracts for the forty nine individuals afflicted with COVID-19, averaging eight per person. For the eight COVID-negative patients in the studied dataset, 121 extract images were collected, averaging fifteen per person. The image extracts are collected rapidly and simply as screen shots from the original CT scan images. The areas selected for screenshots are identified by a radiologist with CT interpretation expertise. The image quality is determined statistically assessing all the pixels in each extracted image.
The greater sampling density for the eight COVID-negative patients is justified to ensure that the image extracts cover a comprehensive range of lung conditions for that group. This is required to provide confidence that a wide range of VS class 0 lung conditions is included in the sample. That is necessary to provide detailed comparisons with all the VS classes of the COVID-positive patients. A potential complication for the analysis conducted is that some of the COVID-negative patients may have, or have had, other lung conditions that have impacted the conditions of their lungs at the time the CT scans were taken. The large age range of the COVID-negative patients and their different lifestyles and home environments are likely to result in these patients displaying a range of lung states as sampled by the CT scans.
A suite of CT scan images taken from an individual patient typically reveals different conditions, reflected by distinct grayscale characteristics, in different portions of the lungs. Indeed, it is not unusual for lung disease sufferers to have substantially more impacts in one lung than the other. Each CT slice and extract images taken from that slice can be assigned a specific VS score reflecting conditions at that point in the patient’s lung. Hence, multiple extract images taken from a suite of CT scans from a single patient are likely to record a range of VS scores. This is a useful outcome, as it enables the CT analyst to pinpoint the position in a sufferer’s lung which is most extensively impacted by the effects of a lung disease, and identify the portions of the lung (or perhaps a specific lung) that are least affected by the disease. As CT images are calibrated on the grayscale of 0 to 255, inter-patient variability should be minimal unless there are calibration issues with a specific CT scan machine, which should be identified by the machine operator.
Five hundred and thirteen quadrilateral CT-image-slice extracts were evaluated in total: one hundred and twenty one from VS class 0; and, three hundred and ninety two from the other four VS classes from COVID-positive patients (53 for VS class 1; 147 for VS class 2; 129 for VS class 3 and 63 for VS class 4). Extract- image dimensions vary between 2000 and 80000 pixels with an average close to 25000 pixels. Extract-image size depends on what is considered to be a representative rectangular area from the pulmonary parenchyma of a left or right lung in a specific CT scan slice. When extracting an image from a CT slice care was taken to limit the area sampled to the parenchyma portion of a lung. This meant preventing the image extracts extending across pleura, diaphragm or mediastinum. As long as the CT‐image extracts are positioned to sample just the parenchyma portion of each lung image, the image extract position, and any image cropping conducted, has minimal impact on the grayscale statistics of the image extract and, therefore, does not affect the performance of prediction models. The highest VS score attained from multiple extract images taken from a single patient is probably the one that is most useful in categorizing the overall severity of lung abnormalities experienced by that patient.
Grayscale images are distinct from bi-tonal black and white images in that they are monochromatic with each pixel possessing a single value that indicates how bright it is on a scale of 0 to 255; where 0 = black, 255 = white and numbers in between 0 and 255 are varying shades of grey. The numerical value assigned to each pixel is in 8-bit integer digital format. The simplicity of the grayscale image structure is conducive for statistical analysis of the distribution of values associated with each pixel in an image. For this study, statistical analysis was implemented with OpenCV software [21] driven by customized Python code. Thirteen statistics were computed for each extract image.
-
Pixel quantity (Pixel#)
-
Average pixel value (on the grayscale 0 to 255)
-
Pixel# displaying the average pixel value
-
Pixel% displaying the average pixel value
-
Variance of pixel values
-
Ratio of variance to average pixel values
-
Standard deviation of pixel values
-
Standard error of the mean pixel values
-
Minimum of the pixel values
-
Tenth percentile (P10) of the pixel values
-
Fiftieth percentile (P50) of the pixel values
-
Ninetieth percentile (P90) of the pixel values
-
Maximum of the pixel values
Pixel# displaying the average pixel value and the standard error (SE = grayscale standard deviation / square root of the quantity of pixels sampled) are statistics that are influenced by image size (i.e., the number of pixels in the specific image analysed). The values of the other statistics, apart from the number of pixels in the image, are independent of image size. SE indicates the degree of uncertainty associated with average grayscale image values. SE is less than 0.7 for all images in the dataset, and, relative to the grayscale of 0–255, that value indicates that there is very low uncertainty associated with the average grayscale value, even for the smallest images. Pixel% displaying the average pixel value, because it is not dependent on image size, is a more useful statistic for comparing a dataset of images. It is therefore used as an input to ML/DL models in preference to the absolute number of pixels associated with the average pixel value.
Machine and Deep Learning Algorithms Applied to Grayscale Statistics
Values of 12 of the 13 statistics derived for each image, omitting the number of pixels in each image, are used as the input variables in this study. The VS class (0–4) assigned to each image by clinical inspection, with respect to the degree of lung abnormalities identified, is the dependent variable that machine learning and deep learning (ML/DL) algorithms attempt to predict from those input variables. The ML/DL algorithms are configured in Python code to solve this classification problem. These algorithms strive to find the minimum root mean squared error (RMSE) of the predicted (VSpred) versus actual (VSact) visual scale assessments, considering all extract images evaluated.
The total of 513 data records (one for each extract image with twelve grayscale statistics and a VS class) are assessed using multiple ML/DL algorithms configured to optimize VS classification. The algorithms applied to this classification task are listed below alphabetically. The detailed methodologies of these algorithms are not presented in detail here as they are all widely used and their methods, as applied to image classification problems, are comprehensively discussed in the literature:
Adaboost (ADA: boosted decision-tree) [22, 23];
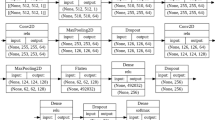
Convolutional Neural Network (CNN; deep learning algorithm) [16, 17];
Decision Tree (DT) [24,25,26];
Extreme Learning Machine (ELM) [27,28,29];
Gaussian Process Classification (GPC; based on the Laplace approximation) [30, 31];
K-nearest Neighbour (KNN) [13, 32];
Multi-layer Perceptron (MLP) [33];
Naïve Bayes Classifier (NBC) [13, 34];
Quadratic Discriminant Analysis (QDA) [35, 36];
Random Forest (RF) [37]; and,
Support Vector Machine (SVM) [38, 39].
That selection includes ten ML algorithms and one DL algorithm (i.e., CNN). This diverse group of algorithms is selected because it covers a wide range of mathematical and logical concepts, and not all of them are dependent on hidden regression and correlation relationships between the variables (e.g., KNN).
Multiple-K‐fold cross‐validation is employed to determine the most statistically reliable divisions of the dataset into training and testing subsets. Four distinct K-folds are considered (fourfold involving 75% training: 25% testing splits; fivefold with 80%: 20% splits; tenfold with 90%: 10% splits; and 15-fold with 93%: 7% splits). Multiple runs are conducted with each K-fold split to generate statistically reliable means and standard deviations of selected error metrics. This method is effective at determining the best splits to use and establishing the uncertainty associated with randomly selected testing subsets. Such analysis is time consuming to conduct when multiple K-folds are considered, so for this study five ML algorithms have been evaluated with multiple-K-fold cross fold analysis (ADA, DT, KNN, RF and SVM). However, the optimum training subset: testing subset split established for these models, for each specific dataset, can be reasonable assumed to be relevant for the other models considered. The multi-K-fold cross validation results obtained from the analysis conducted suggested that a split of 80% training subset: 20% testing subset worked well for the dataset evaluated and this division was randomly applied for this study.
Each of the ML/DL models requires tuning of the hyperparameters (control values) to be applied for each specific dataset to which they are applied. This involves finding the optimum values that minimize prediction errors associated with each model. It requires multiple sensitivity test runs being conducted for each ML/DL model, each applying different potentially feasible control values. This optimization has been achieved for this study using a combination of trial-and-error analysis, grid search and Bayesian optimization, making use of the available Scikit learn functions to perform the latter two sensitivities. The optimized hyperparameters adopted for each algorithm are described in Table 1.
Metrics Used to Assess the Accuracy of ML/DL Predictions
Several commonly used statistical measures of prediction accuracy (Fig. 3) are used to assess the prediction accuracy achieved by the ML/DL algorithms. These accuracy assessment metrics are useful to consider collectively when comparing the prediction accuracy achieved by particular algorithms. However, MSE and RMSE are, to some extent, more pertinent accuracy measures as these are the values that the algorithms are trying to minimize as an objective function.
Results
Grayscale Image Statistics
The value distributions of the statistical attributes assessed for the CT scan extract images are summarized in Table 2. The table is divided to consider the dataset as a whole (513 data records), the 121 images from COVID-negative patients and the 392 images from the COVID-positive patients. Several of these grayscale statistical attributes display a substantial range of values. These distributions are illustrated in Fig. 4 with box and whisker diagrams, distinguishing the COVID-negative and COVID-positive image sample sets for each grayscale attribute. By juxtaposing the box and whisker diagrams for each attribute for those two sample sets, plotted on the sample scale range, the differences are clear to see. The average grayscale, variance grayscale, P10/P50/P90 grayscale and pixel% at the average grayscale, in particular, show quite distinctive distributions for those individual afflicted with COVID-19 and those unaffected by it (Table 2, Fig. 4). Such differences form the basis for using them to discriminate between the impacts related to pulmonary conditions such as COVID-19.
Box/whisker distribution plots of CT image extract grayscale attributes. For each attribute the distribution of the COVID −ve samples (blue boxes) is placed beside the distribution of COVID + ve samples (red boxes), displayed on the same scale range. In each plot the boxes express the range of the second and third distributions quartiles; crosses within the boxes represent the distribution means; horizontal lines in each box represent the distribution medians; the vertical lines and whiskers express the confident limits of the distributions; and the dots outside the whiskers represent potential outliers positioned beyond ± 1.5 times the interquartile range from the boxes
Correlation among the grayscale statistical attributes and with VS are also encouraging (Table 3). There are strong positive Pearson correlation coefficients (R) with VS displayed by grayscale P10, P50, Average, P90 and variance. A strong negative R value exists between VS and the percent of pixels at the grayscale average. These correlations indicate that several of the grayscale statistical attribute distributions are varying systematically across the image extract dataset (Table 2; Fig. 4) between COVID-19 −ve and COVID-19 + ve samples. In addition to R, the Spearman rank correlation coefficient (p) values are also displayed to express the correlation relationships between VS and the grayscale statistical attributes. A key assumption of R is that the variable distributions being correlated are normally (symmetrically) distributed, that is they are parametric in their behaviour. On the other hand, p makes no such assumptions and is non-parametric because it is calculated using ranking positions rather than absolute data values.
In general, the R and p values are quite close (last two columns in Table 3) implying that the grayscale statistical attributes and VS distributions are not highly skewed and that they are not highly non-parametric. The grayscale P90 displays the strongest positive R (0.87) and p (0.88) correlations with VS. The average pixel (grayscale) value also displays strong positive R (0.78) and p (0.81) correlations with the dependent variable (VS). The P50 pixel value R (0.72) and p (0.75) correlations with VS are only slightly less strong. Grayscale variance values also display a strong positive R (0.67) and p (0.72) correlations with VS. Pixel% at the average value displays robust negative R (− 0.75) and p (− 0.80) correlations with VS. These relationships suggest that the grayscale statistical measures, particular those displaying high correlation coefficients with VS, are likely to be exploitable by ML/DL methods to accurately predict VS.
Relationships Between Grayscale Statistical Attributes and VS
Figures 5, 6 and 7 graphical express the continuity and extent of the key grayscale statistical attributes for the CT-slice-extract images displaying VS and the severity of pulmonary impacts. Figure 4 displays the scaled relationships between the distributions of average pixel value versus variance of the pixel values versus pixel% at the average value. The VS values are distinguished for each extract image in Fig. 5. Scale factors are used for variance values and pixel% at the average value to centralize the data point distributions within the triangular display (Fig. 5).
Three grayscale statistical attributes possessing high correlation coefficients with VS (Table 3) plotted in a scaled triangular diagram for the purpose of distinguishing the severity of lung abnormalities. Grp 0 = COVID-negative; Grp 1–4 represent increasing severity of lung abnormality in COVID-positive patients
The average value, variance value and pixel% at the average value can, to a degree, distinguish the extent of pulmonary impacts related to COVID-19 (Fig. 5). Strikingly, there is a continuous progression from VS class 0 (lower-left portion of triangle) to VS class 4 (middle-to-upper-right portion of triangle). VS class 0 and VS class 3 are clearly separated on this display. However, there is substantial overlap between VS Classes 1, 2 and 4 and the other VS classes using this plot in isolation. The trend and distribution of data records in Fig. 5 highlight that lungs with no, or trace, abnormalities are characterized by low grayscale average and variance. Progressively, average values and variance values increase and pixel% at the average values decreases as lung abnormalities become more substantial. Those lungs associated with the most severe abnormalities approach right-side triangular apex (Fig. 5). This is because the grayscale in such images is dominated by light-grey shades causing grayscale variance to decrease and grayscale average to increase substantially in lungs with such severe abnormalities. The progressive trend is therefore initially from southwest to northeast (VS class 0 to VS class 3) in Fig. 5, and then from northeast to southeast (VS class 3 to VS class 4).
Three-dimensional (3D) graphics are also useful for displaying the relationships between the grayscale statistical attributes (e.g., Figs. 6 and 7). Grayscale P90 replaces grayscale average from Fig. 5 to provide the 3-D display shown in Fig. 6. There is a clear progression from lower right (VS class 0) to top left (VS class 4) in Fig. 6, although, as with Fig. 5, a degree of overlap exists among the VS classes. In Fig. 7 (P10, average and P90 grayscale statistics) there is a progression from lower left (VS class 0) to top right (VS class 4), combined with a degree of overlap between the VS classes.
Grayscale statistical distributions (Tables 1 and 2; Figs. 5, 6 and 7) in extract images from CT scans clearly offer the potential to distinguish the severity of pulmonary impacts in those individuals afflicted with COVID-19. Although there are several statistical attributes displaying high correlation coefficients with VS and combinations of them can achieve good distinctions between certain VS classes (Figs. 5, 6 and 7), the overlap between certain classes makes such 3-D graphics unsuitable for definitive predictions of VS. This limitation justifies the deployment of ML/DL algorithms to consider all of the grayscale statistical attributes associated with the CT extract images to see if it is possible to predict and distinguish all classes of VS with a higher degree of confidence. By including several additional grayscale statistic attributes with relatively low correlation coefficients with VS (Table 3), the ML/DL algorithms are able to exploit more subtle relationships between them to provide better VS predictions.
Selecting Features for Sensitivity Analysis
The statistical (grayscale) attribute dataset of CT scan extract images is comprised of 513 data records. It includes 13 independent variables and one dependent variable (Tables 2). The 13 input variables are all available for use by the ML/DL algorithms for VS prediction. Some sensitivity testing was conducted, based on the relative influence of the variables to identify which of these grayscale statistics could be omitted without reducing the VS prediction accuracy of the models. Models considering just nine of the independent variables (i.e., leaving out pixel#, standard error of the mean, minimum value and variance/average ratio), 10 variables (as for the 9-variable case but including minimum grayscale), 11 variables (as for the 10-variable case but including standard error) and 12 variables (as for the eleven-variable case but including variance/average grayscale ratio) were evaluated with the ML/DL models. All of those cases generated credible predictions with high accuracy. However, the 12-variable case outperformed the other cases, demonstrating that all those variables are able to make useful contributions to VS prediction. Consequently, it was the 12-variable case (excluding the number of pixels per image) that was selected for detailed analysis.
Performance Comparison of ML/DL Methods for Predicting VS
Table 4 presents the MAE result of fivefold cross validation analysis applied to selected ML models configured to predict VS. Multiple K-fold analysis was conducted (fourfold, fivefold, tenfold and 15-fold) but the fivefold analysis generated the most consistent results. These results justify the use of a 80% training subset: 20% testing subset split. The fivefold results involve fifteen separate random 80%:20% data record splits, presenting the MAE means and standard deviations for the 20% testing subsets (Table 4). It is apparent that the RF and KNN models generate statistically lower errors than the other ML models evaluated.
Table 5 lists the accuracy of predicting (VSpred versus VSact) as determined by the statistical measures defined in Fig. 3 for the eleven ML/DL algorithms. These measures provide a comparison of each model’s capability to correctly predict /classify the VS value of all five hundred and thirteen data records in the image extract dataset on a supervised learning basis. In addition to the prediction accuracy measures, each algorithm is ranked in Table 5 in ascending order of the RMSE values achieved and the quantity of errors made in its predictions (the two right-side columns in Table 5).
The CNN deep learning model outperforms all the ML models resulting in just 18 VS prediction errors (out of a possible 513), achieving an RMSE of 0.19 and R2 of 0.98. Of the ML models, RF delivers the most accurate results (26 prediction errors, RMSE of 0.26 and R2 of 0.96) and the ADA, KNN, DT and ELM models also achieve impressive accuracy. The MLP, NBC and QDA algorithms substantially underperform in terms of their VS prediction accuracy. The variations in APD and AAPD (Table 5) are broadly consistent with those for MAE and RMSE, reinforcing the performance order of the classification models. Careful checking of the images that are predicted incorrectly by the different ML/DL models evaluated reveals that there is no definitive difference between them and the correctly predicted images. A small number of such errors are to be expected as the VS classes grade into each other and overlap to an extent depending upon the combination of grayscale attributes considered (e.g. Fig. 5).
It is notable that some algorithms rank differently based on RMSE performance compared to the numbers of prediction errors generated. For instance, ELM ranks 4 in terms of RMSE but ranks 6 based on the number of errors generated (Table 5). On the other hand, ADA ranks 5 in terms of RMSE but ranks 4 on the basis of error numbers. The reason for this is due to the magnitude of the prediction errors made. If an algorithm makes a prediction error by placing the data record in an adjacent class that will have a smaller impact on increasing its RMSE than if the error places the data record two of more classes away from the correct class. It is clearly important to consider both types of error. Confusion matrices help to identify the reliability of the algorithms in terms of the relative severity of the errors they make.
Confusion Matrices to Assess VS Prediction Performance
Although the ML/DL algorithms configured to minimize RMSE (i.e. RMSE is their objective function) the ultimate objective of the VS prediction effort is to minimize the number of data records that are incorrectly predicted. It is useful to configure the algorithms in this way because RMSE provides a continuous scale which the algorithms can progressively minimize. It is possible for the number of errors to move up and the RMSE value to move down; compare, for example, the outcomes for the ADA and ELM algorithms in Table 5.
A confusion matrix provides a detailed analysis of the nature of the misclassifications made by specific algorithms. These diagrams provide more detail about how each prediction model is performing. They identify the VS classes that an algorithm predicts more accurately than others. They also identify which classes an algorithm is most prone to confuse. Figure 8 displays three such confusion matrices for the CNN, KNN and ADA models.
It is apparent from Fig. 8 that the VS prediction models perform quite differently in their ability to predict specific VS classes. The best-performing CNN model (Fig. 8a) is most accurate when making class 0 predictions and least accurate when making class 1 predictions. Overall, in percentage terms it predicts classes 0 to 2 with higher accuracy than classes 3 and 4. Of note, is that the CNN model does not involve prediction errors that are placed greater than a single VS class from the actual VS value. This feature, combined with fewer errors (18) explains why it can be considered as the most reliable VS class predictor of the models evaluated.
Figures 8b and 8c show confusion matrices for the KNN and ADA models, respectively. These are two high-performing ML models that both generate 28 total errors. However, the distribution of the prediction errors is quite different for each of these models. The KNN results involve 5 errors that are more than one class removed from the actual VS class. On the other hand the ADA results involve 8 errors that are more than one class removed from the actual VS class. This explains the RMSE values achieved by the two models: KNN = 0.2887; ADA = 0.3175. Also of interest is that both models are most likely to confuse VS class 2 (11 errors each; nearly 40% of their total errors). On the other hand, KNN is most reliable in its predictions of class 0 and 1. Indeed, the KNN model outperforms the CNN model in its class 1 prediction performance. In contrast, ADA performs less well in predicting class 0, confusing 5 images as VS class 1 and one image as class 2. However, the ADA model shows better prediction performance for VS class 4 than either the CNN or KNN models.
By highlighting which VS classes are predicted most reliably by each ML/DL model, the confusion matrices provide the analyst with the ability to select prediction models that best suit tasks focused on distinguishing specific VS classes. Analysis of confusion matrices for these class prediction models therefore usefully complements the error/accuracy statistical measures established for each ML/DL model. It also suggests that running an ensemble of several models is advisable as each model’s prediction accuracy varies in its ability to accurately predict specific VS classes.
Simplified Statistical Scoring System Using Selected Grayscale Statistics
The ranges displayed by the grayscale statistical attribute variables (see “Relationships between grayscale statistical attributes and VS”) suggest relatively basic formulaic relationships among just a few of these variables could predict the severity of lung abnormalities from CT scan image extracts to a reasonable level of accuracy. The accuracy achievable would clearly be less than that demonstrated for the ML/DL models in relation to the VS classes. However, this could provide the basis for developing an objective, automated algorithmic scale of severity associated with lung abnormalities from CT extract images. Such an automated scale could be used to complement the VS score assigned by a clinician (i.e., involving human interpretation).
It is feasible to create a simplified lung abnormality statistical scoring system based on algorithmic relationships involving just a few of the most influential grayscale statistics with respect to VS. Such an algorithmic scoring (AS) approach is useful to compare with the visual assessments and potentially offers a means to provisionally automate CT scan assessments prior to expert visual assessment. An example of one such AS system is provided and assessed. It involves just five of the grayscale statistics recorded (P10, average, P90, variance and pixel% at the average value), i.e., those showing distinctive and progressive separations in Figs. 5 and 6 and high correlation coefficients with VS.
There are just four groups (1 to 4) in the AS system described. Unlike the VS with five classes, there is no AS group 0 representing COVID negative patients. AS just focuses on the degree of lung abnormalities, with group 1 at the low-lung-abnormality end of the scale and group 4 at the high end. There is no attempt made in AS to distinguish COVID negative patients from those COVID positive patients displaying no discernible lung abnormalities. This means that most images that fall into classes 1 and 2 of the VS system would be expected to fall into AS group 1.
The algorithmic rules and logical sequence used to assign the images to specific groups for the AS groups are as follows:
-
A.
AS Class 4 (severe category) is distinguished first at the high end of the scale on the basis that images must exceed all of these three statistical limits: P10 grayscale > = 100, Average grayscale > = 150 and P90 grayscale > = 200.
-
B.
AS Class 1 (normal/minimal lung abnormalities) is then distinguished at the low end of the scale on the basis that images must fall within these four statistical limits: P10 grayscale < 80, P90 grayscale < 125, variance < 1000 and pixel% at the average value > 1.5%.
-
C.
AS Class 2 (minor lung abnormalities) is then distinguished for those images that have not already been allocated to AS classes 1 or 4 by applying two statistical limits: average grayscale < 125 and P90 grayscale < 150.
-
D.
AS Class 3 (substantial lung abnormalities) is assigned to those images that do not fall within the limits specified for AS classes 1,2, and 4.
Figure 9 displays the 513 images assessed with this AS system. The numbers of images assigned to each AS class are: AS class 1 = 116; AS class 2 = 117; AS class 3 = 236; and AS class 4 = 44. It is clear from Fig. 9 that there is much greater separation between the AS classes than the VS classes (Fig. 5) for the image extracts considered. This makes it more suitable for a quick-look, consistent diagnosis based on relatively few (just 5) grayscale statistical variables, removing the potential subjectivity of expert (human) visual assessments.
Triangular display of key grayscale statistics for distinguishing severity of lung abnormalities with the algorithmic statistical scoring classification system applied. The AS scoring scale consists of just four classes (group 1 to group 4). The segregation of the groups is more apparent than using the visual scoring (VS) system (Fig. 5) in which there is substantially more overlap between the groups
The relatively simple segmentation criteria for the AS classes does not, however, consider all the grayscale statistical information recorded in the grayscale analysis of each image (Table 1). It is useful therefore to evaluate whether ML/DL algorithms can more accurately predict the AS groups than the VS classes for this dataset.
Table 6 presents the MAE results of fivefold cross validation analysis applied to selected ML models configured to predict AS. Multiple K-fold analysis was conducted (fourfold, fivefold, tenfold and 15-fold) but the fivefold analysis generated the most consistent results. These results justify the use of a 80% training subset: 20% testing subset split for the AS predictions. The fivefold results involving fifteen separate random 80%:20% data record splits, presenting the MAE means and standard deviations for the 20% testing subsets (Table 6). It is apparent that the RF, DT and ADA models generate statistically lower errors than the other ML models evaluated.
Table 7 displays the results of the ML/DL models applied (with the same control parameters as used for the VS predictions, Table 3) to the entire 513 data records using the 12 grayscale statistical variables. It demonstrates that the AS system is easier for the ML/DL algorithms to distinguish than the VS system and the best-performing algorithms achieve higher prediction accuracy with much fewer errors.
Impressively, the best performing ML algorithms (RF and DT) achieve AS prediction accuracy of > 99% with only one confused predictions out of the 513 images assessed. This outperforms the CNN deep learning model, which is actually ranked sixth in terms of its performance compared to other algorithms. Inspection of the confusion matrices reveals that DT makes its one prediction error, confusing an AS class 2 image as class 1. On the other hand, the RF model makes its one prediction error confusing an AS class-4 image as an AS class-3 image. Cross-validation analysis confirms that the ML/DL models as configured do not overfit AS dataset (Table 6). The reason why there are much fewer prediction errors generated by the models when applied to the AS dataset is that the classification methodology distinguishing the AS classes is based on a simplified statistical scale compared to the VS dataset. The AS classification does not depend on clinicians’ judgement but is determined solely by a formulaic algorithm. Consequently, the AS dataset shows less overlap between its classes than the VS dataset, as revealed by comparing Fig. 5 (VS dataset) with Fig. 9.
It is expected that other algorithmic combinations of image grayscale statistical attributes may be able to match or exceed the performance of the simple AS method described here. Further research is required to verify this. However, the AS described demonstrates the viability of an algorithmically derived, severity of lung abnormality scale based on CT image extract grayscale statistical attributes.
Discussion
Over the past 2 years many ML and DL models have been developed and evaluated to assess CT lung scan images to determine whether a patient is, or is not, suffering from COVID-19. A recent list of deep learning models and the accuracies they achieve is provided by Garg et al. [40]. Most of these studies address binary classification analysis (COVID-positive versus COVID-negative) although some [40, 41] do distinguish three classes (in addition distinguishing those images related to patients suffering other lung diseases). Most of these deep learning models use activation maps of an entire CT images to make their classifications. This study is unique in that it aims to not only distinguish COVID-positive from COVID-negative patients but also make distinctions between degrees of severity of lung abnormalities. Moreover, the method proposed is more transparent about the features used to influence its class selections. Many of the binary- and tertiary-class-selection, deep learning models proposed are not very transparent concerning the specific image criteria used to make their class selections, other than revealing different weights assigned to different image manipulation functions.
Analysis presented here for both the VS and AS approaches to classifying the degree of pulmonary impacts in COVID-19 patients provide sufficient encouragement to justify more extensive future research with respect to grayscale statistical attributes of image extracts taken from CT scan slices. Indeed, the approach may also be worth evaluating for the assessment of other lung diseases using CT-image data. The extensive value ranges of several of these grayscale statistical attribute distributions (Table 2, Fig. 4), and the correlation relationships between them (Table 3), are conducive to beneficial exploitation by algorithmic relationships and/or ML/DL models to grade and quantify the severity of lung abnormalities quite precisely. In particular, the average grayscale, variance grayscale, P10/P50/P90 grayscale and pixel% at the average grayscale, can collectively be used, to an extent, to distinguish between those individuals afflicted with COVID-19 and those unaffected by it. However, the leave-one-out analysis conducted as part of feature selection, for the ML/DL model development, indicates that twelve of the attributes (all of those listed in Table 2 except number of pixels), when used collectively, lead to the lowest classification errors. Hence, some of the attributes with relatively low correlation coefficients with the VS classes or AS groups do make useful contributions to the ML/DL class predictions.
Graphic representations (Figs. 5, 6, 7 and 9) of selected attributes are informative for quick-look assessments of the CT scan extract images. However, for more reliable classification of the images to either the VS or an AS system, ML/DL methods, deployed on a supervised learning basis, are required. The best of these (CNN) achieves better than 96% prediction accuracy (and R2 > 0.98) for the VS classification applied to the entire five hundred and thirteen image dataset evaluated. Moreover, the DT and RF algorithms achieve 98.8% prediction accuracy (and R2 = 0.998), with just 1 prediction error in the five hundred and thirteen images classified, for the AS classification system for the same image dataset. The future research planned will address applying the methods to larger datasets of CT scan slices, evaluating alternative and more complex algorithmic AS logic, and automating the technique with image segregation software for rapid evaluation of a broader range of extract image shapes and/or extract image compilations from multiple slices. There are now some open-access chest CT image repository datasets available [42, 43] that make such expanded studies possible. Until such work is completed, questions remain concerning the generalizability of the method beyond the dataset evaluated in this study.
Clearly, the VS systems benefit from the clinical expertise of visual observation involving a human being. On the plus side. clinical experts are able to use a broader range of factors than those available from grayscale statistical analysis alone. On the downside, the VS class boundaries are associated with a degree of subjectivity potentially varying slightly from the inspection of one clinician to another. Figures 5, 6, 7 highlight that the severity of lung abnormalities extends over a broad and continuous spectrum of image attributes. Those grayscale statistical attribute values are not conveniently segregated into clusters which might improve the definitions of the class boundaries. Consequently, any class boundaries, either visually or algorithmically defined, will be arbitrarily placed within this continuous spectrum of grayscale attribute values. Taking the arbitrary nature of the placement of the VS class boundaries into account, it is impressive that the CNN method, on a supervised learning basis, can accurately predict the VS classes resulting in just eighteen prediction errors from five hundred and thirteen image extracts evaluated. It seems possible with larger datasets that the deep learning models should be able to approach zero VS prediction errors on a supervised basis and achieve high VS class prediction accuracy on a semi-supervised basis. Indeed, for the AS approach the best ML models have achieved just one AS group prediction error from the data set evaluated (Table 7 and Fig. 9).
The low prediction errors (high classification accuracy) generated by the best performing ML/DL models applied in the two dataset configurations studied, VS (96.5% accuracy) and AS (99.8% accuracy), compare well with the results of other studies. This is particularly impressive because other published ML/DL studies focus their learning models on just the binary classification of distinguishing COVID-19 negative from COVID-19 positive images, not on the five class (VS dataset) and four class (AS dataset) classifications of severity of lung condition attempted by this study. One published study classified one hundred CT-scan images with a CNN model with 85% accuracy in that binary classification task [44]. Binary classification accuracy was improved upon to reach an accuracy 98% with another eight hundred and twelve CT-scan dataset [45]. Also, applying DL models to the binary classification of three hundred and sixty CT-scan images, another study reported 91% classification accuracy [46]. Two further DL studies based on several thousand X-ray images [47] and CT-scan images [48] achieved 73% and 82% binary classification accuracy, respectively”.
For the VS class predictions, it is particularly impressive that several of the ML/DL algorithms are able to distinguish with high accuracy between VS class 0 (COVID-free) and VS class1 (COVID-afflicted but displaying negligible or trace image indications of pulmonary impacts. Consider, for example, the performances of the CNN and KNN models (Fig. 8). Clinical experts find it extremely difficult to separate VS classes 0 and 1, solely by visual analysis of the CT images, with any degree of accuracy (i.e., in the absence of a rRT-PCR COVID test). Yet with > 95% accuracy (Fig. 8a and b) the CNN and KNN 12-variable models can correctly discriminate images between these two VS classes. In the case of the KNN model, just 3 errors are generated from the 174 images that belong to VS classes 0 and 1. This result confirms that the grayscale image statistical attributes are capable of distinguishing facets from these images that are not readily discernible by human visual inspection, even by an expert clinician. This capability demonstrates the wealth of information that can be gained from CT scan slice extract images using ML/DL techniques applied to the grayscale statistical attribute distributions they contain.
Conclusions
Grayscale statistical analysis accurately predicts visual assessments of CT scans made by clinicians that assign each image a visual score (VS) on the scale 0 to 4. VS class 0 refers to images from individuals not afflicted with COVID-19. VS class1 refers to those individuals afflicted with COVID-19 but without (or with only trace) pulmonary impacts. VS classes 2 to 4 refer to increasing degrees of pulmonary impacts in individuals afflicted with COVID-19. Standalone graphical analysis of the grayscale image statistical attributes showing the strongest correlations with VS is insightful but unable to definitively predict VS classes. However, evaluation of eleven machine learning and deep learning (ML/DL) models with twelve image grayscale statistical attributes as input variables, demonstrates that VS class prediction can be achieved with up to 96.5% accuracy (R2 = 98.05; just eighteen out of five hundred and thirteen images incorrectly classified) for the best performing convolutional neural network (CNN) model.
Additionally, the image grayscale statistics can be combined to derive an automated algorithmic scoring (AS) systems based on just a few of the attributes showing high correlation coefficients with VS. One such AS system, based on just five attributes (grayscale P10, average, P90, variance and the pixel% at the average value) can formulaically define a four-group AS scale. That AS scale can be assessed graphically to make reasonable group distinctions. However, when that AS scale is evaluated with twelve grayscale image attributes using ML/DL models it provides much improved group prediction accuracy. The AS scale defined varies from AS group 1 with no or trace lung abnormalities, for patients with and without COVID-19, up to AS group 4 for patients with severe lung abnormalities. Decision tree (DT) and random forest (RF) ML models manage to predict the AS group for CT scan image extracts with 99.8% accuracy (R2 = 99.8; just one out of five hundred and thirteen images incorrectly classified) using 12 grayscale statistical attributes as input variables. The CNN deep learning model is outperformed by several ML models (RF, DT, Adaboost, Gaussian process classification and K-nearest neighbour) in the prediction of the AS scale, although it still manages to achieve 98.4% AS prediction accuracy (R2 = 98.2; eight out of five hundred and thirteen images incorrectly classified).
These results are very encouraging regarding the potential for using image extract grayscale statistics to develop rapid and precise expert systems for predicting the severity of lung abnormalities from pulmonary CT scan slices. The prediction errors of the ML/DL models for both VS and AS scales, analysed with the aid of confusion matrices, reveal details of the relative capabilities of the models to distinguish individual VS classes and AS groups. With respect to the VS scale, confusion-matrix diagrams identify the ML/DL algorithms best suited to distinguish VS class 0 (individuals without COVID-19) from VS class 1 (individuals with COVID-19 but showing negligible or only trace, visually discernible, abnormalities). These algorithms are able to do this with very few prediction errors. For example, the KNN model delivered just three misclassifications out of 174 images belonging to VS classes 0 and 1. What is impressive about that prediction performance is that clinical experts struggle to make the distinction between the classes 0 and 1 (as defined) by visual analysis of the CT-scan data in isolation, i.e., without the availability of a rRT-PCR laboratory test result to guide them. Further studies are required using larger datasets to assess the generalizability of the method beyond the dataset evaluated in this study, and to potentially automate the grayscale image extraction process.
Data availability
There is no shared data available with this study.
References
Bai H, Hsieh B, Xiong Z, Halsey C, Choi JW, et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020;296(2):E46–54. https://doi.org/10.1148/radiol.2020200823.
Hare SS, Tavare AN, Dattani V, Musaddaq B, Beal I, et al. Validation of the British Society of Thoracic Imaging guidelines for COVID-19 chest radiograph reporting. Clin Radiol. 2020;75(9):710.e9-710.e14. https://doi.org/10.1016/j.crad.2020.06.005.
Ai T, Yang Z, Hou H, Zhan C, Chen C, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–40. https://doi.org/10.1148/radiol.2020200642.
Fang Y, Zhang H, Xie J, Lin M, Ying L, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–7. https://doi.org/10.1148/radiol.2020200432.
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:200343. https://doi.org/10.1148/radiol.2020200343.
Ng MY, Lee EY, Yang J, Yang F, Li X, et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiology. 2020;2(1):e200034. https://doi.org/10.1148/ryct.2020200034.
Li K, Wu J, Wu F, Guo D, Chen L, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55(6):327–31. https://doi.org/10.1097/RLI.0000000000000672.
Farhat H, Sakr GE, Kilany R. Deep learning applications in pulmonary medical imaging: recent updates and insights on COVID-19. Mach Vis Appl. 2020. https://doi.org/10.1007/s00138-020-01101-5.
López-Cabrera JD, Orozco-Morales R, Portal-Diaz JA, Lovelle-Enríquez O, Pérez-Díaz M. Current limitations to identify COVID-19 using artificial intelligence with chest X-ray imaging. Heal Technol. 2021;11(2):411–24.
Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology. 2020;295:202–7. https://doi.org/10.1148/radiol.2020200230.
Duzgun SA, Durhan G, Demirkazik FB, Ariyurek AMG, OM,. COVID-19 pneumonia: the great radiological mimicker. Insights Imaging. 2020;11:118. https://doi.org/10.1186/s13244-020-00933-z.
Yu Z, Li X, Sun H, Wang J, Zhao T, et al. Rapid identification of COVID-19 severity in CT scans through classification of deep features. BioMed Eng OnLine. 2020;19:63. https://doi.org/10.1186/s12938-020-00807-x.
Hussain L, Nguyen T, Li H, Abbasi AA, Lone KJ, Zhao Z, et al. Machine-learning classification of texture features of portable chest X-ray accurately classifies COVID-19 lung infection. BioMed Eng OnLine. 2020;19:88. https://doi.org/10.1186/s12938-020-00831-x.
Shi F, Wang J, Shi J, Wu Z, Wang Q, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2021;14:4–15. https://doi.org/10.1109/RBME.2020.2987975.
Dong D, Tang Z, Wang S, Hui H, Gong L, et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng. 2021;14:16–29. https://doi.org/10.1109/RBME.2020.2990959.
Li L, Qin L, Xu Z, Yin Y, Wang X, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–71. https://doi.org/10.1148/radiol.2020200905.
Song Y, Zheng S, Li L, Zhang X, Zhang X, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans Comput Biol Bioinform. 2021. https://doi.org/10.1109/TCBB.2021.3065361.
Haque IRI, Neubert J. Deep learning approaches to biomedical image segmentation. Inform Med Unlocked. 2020;18:100297. https://doi.org/10.1016/j.imu.2020.100297.
Zhou T, Canu S, Ruan S. An automatic COVID-19 CT segmentation based on U-Net with integrated spatial and channel attention mechanism. Int J Imaging Syst Technol. 2021;31:16–27. https://doi.org/10.1002/ima.22527.
Amber Diagnostics. Different types of CT machines [Accessed 14 September, 2021] https://www.amberusa.com/blog/types-of-ct-machines/. 2021
OpenCV. Open source computer vision library. [Accessed 14 September 2021] https://docs.opencv.org/master/d1/dfb/intro.html. 2021
Barstugan M, Rahime Ceylan R, Sivri M, Erdogan H. Automatic liver segmentation in abdomen CT images using SLIC and AdaBoost algorithms. In: Proceedings of the 2018 8th International Conference on Bioscience, Biochemistry and Bioinformatics 129–133. 2018. https://doi.org/10.1145/3180382.3180383
Kuwahara, M, Kido S, Shouno H. Classification of patterns for diffuse lung diseases in thoracic CT images by AdaBoost algorithm. In: Proceedings Volume 7260, Medical Imaging: Computer-Aided Diagnosis 726037. 2009. https://doi.org/10.1117/12.811497
Agarwal C, Sharma A. Image understanding using decision tree based machine learning. In: Proceedings of the 5th international Conference on Information Technology & Multimedia. 2011. https://doi.org/10.1109/ICIMU.2011.6122757
Rajendran P, Madheswaran M. Hybrid medical image classification using association rule mining with decision tree algorithm. J Comput. 2020;2(1):127–36.
Yoo SH, Geng H, Chiu TL, Yu SK, Cho DC, et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front Med. 2020;7:427. https://doi.org/10.3389/fmed.2020.00427.
Albadr MAA, Tiun S, Ayob M, Al-Dhief FT, Omar K, Hamzah FA. Optimised genetic algorithm-extreme learning machine approach for automatic COVID-19 detection. PLoS ONE. 2020;15(12):e0242899. https://doi.org/10.1371/journal.pone.0242899.
Dogantakin A, Ozyurt F, Avci E, Koc M. A novel approach for liver image classification: PH-C-ELM. Measurement. 2019;137:332–8. https://doi.org/10.1016/j.measurement.2019.01.060.
Huang GB, Zhou H, Ding X, Zhang R. Extreme learning machine for regression and multiclass classification. IEEE Trans Syst Man Cybern Part B (Cybern). 2011;42:513–29. https://doi.org/10.1109/TSMCB.2011.2168604. (PMID: 21984515).
Zhu F, Carpenter T, Gonzalez DR, Atkinson M, Wardlaw J. Computed tomography perfusion imaging denoising using gaussian process regression. Phys Med Biol. 2012;57(12):N183-198. https://doi.org/10.1088/0031-9155/57/12/N183.
Hussein S, Gillies R, Cao K, Song Q, Bagci U. TumorNet: Lung nodule characterization using multi-view Convolutional Neural Network with Gaussian Process. In: IEEE 14th International Symposium on Biomedical Imaging. 2017. https://doi.org/10.1109/ISBI.2017.7950686
Ramteke RJ, KhachaneMonali Y. Automatic medical image classification and abnormality detection using K-nearest neighbour. Int J Adv Comput Res. 2012;2(4–6):190–6.
Kuruvilla J, Gunavathi K. Lung cancer classification using neural networks for CT images. Comput Methods Prog Biomed. 2014;113(1):202–9. https://doi.org/10.1016/j.cmpb.2013.10.011.
Park DC. Image classification using naïve Bayes classifier. Int J Comput Sci Electron Eng. 2016;4(3):135–9.
Suzuki K. Machine learning in computer-aided diagnosis of the thorax and colon in CT: a survey. IEICE Trans Inf Syst. 2013;E96(D4):772–83. https://doi.org/10.1587/transinf.e96.d.772.
Huidrom R, Chanu YJ, Singh KM. Pulmonary nodule detection on computed tomography using neuro-evolutionary scheme. SIViP. 2019;13:53–60. https://doi.org/10.1007/s11760-018-1327-4.
Nedjar I, El Habib DM, Settouti N. Random forest based classification of medical X-ray images using a genetic algorithm for feature selection. J Mech Med Biol. 2015;15(2):1540025. https://doi.org/10.1142/S0219519415400254.
Cyran KA, Kawulok J, Kawulok M, Stawarz M, Michalak M, et al (2012) Support vector machines in biomedical and biometrical applications. In: Ramanna S, Jain LC, Howlett RJ (eds) Emerging paradigms in ML. smart innovation, systems and technologies, 13. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-28699-5_15
Zhang Z, Sejdic E. Radiological images and machine learning: Trends, perspectives, and prospects. Comput Biol Med. 2019;108:354–70. https://doi.org/10.1016/j.compbiomed.2019.02.017.
Garg A, Salehi S, La Rocca M, Garner R, Duncan D. Efficient and visualizable convolutional neural networks for COVID-19 classification using Chest CT. Expert Syst Appl. 2022;195:116540. https://doi.org/10.1016/j.eswa.2022.116540.
Baghdadi NA, Malki A, Abdelaliem SF, Balaha HM, Badawy M, Elhosseini M. An automated diagnosis and classification of COVID-19 from chest CT images using a transfer learning-based convolutional neural network. Comput Biol Med. 2022;144:105383. https://doi.org/10.1016/j.compbiomed.2022.105383.
Shakouri S, Bakhshali MA, Layegh P, Kiani B, Masoumi F, AtaeiNakhaei S, Mostafavi SM. COVID-19-CT-dataset: an open-access chest CT image repository of 1000+ patients with confirmed COVID-19 diagnosis. BMC Res Notes. 2021;14(1):3. https://doi.org/10.1186/s13104-021-05592-x.
Zhao J, Zhang Y, He X, Xie P. Covid-CT-dataset: a CT scan dataset about COVID-19. 2020. https://doi.org/10.48550/arXiv.2003.13865. arXiv preprint arXiv:2003.13865v3 [cs.LG]
Polsinelli M, Cinque L, Placidi G. A light CNN for detecting COVID-19 from CT scans of the chest. Pattern Recogn Lett. 2020;140:95–100. https://doi.org/10.1016/j.patrec.2020.10.00.
Arora V, Ng EY, Leekh RS, Darshan M, Singh A. Transfer learning-based approach for detecting COVID-19 ailment in lung CT scan. Comput Biol Med. 2021;135:104575. https://doi.org/10.1016/j.compbiomed.2021.104575.
Dansana D, Kumar R, Bhattacharjee A, Hemanth DJ, Gupta D, Khanna A, Castillo O. Early diagnosis of COVID-19-affected patients based on X-ray and computed tomography images using deep learning algorithm. Soft Comput. 2020. https://doi.org/10.1007/s00500-020-05275-y.
Bharati S, Podder P, Rubaiyat M, Mondal H. Hybrid deep learning for detecting lung diseases from X-ray images. Inform Med Unlocked. 2020;20:100391. https://doi.org/10.1016/j.imu.2020.100391.
Bharati S, Podder P, Mondal MRH, Gandhi N (2021). Optimized NASNet for diagnosis of COVID-19 from lung CT images. In: Abraham A, Piuri V, Gandhi N, Siarry P, Kaklauskas A, Madureira A (eds) Intelligent systems design and applications. ISDA 2020. Advances in intelligent systems and computing, 1351. https://doi.org/10.1007/978-3-030-71187-0_59.
Funding
No external funding was received in association with this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghashghaei, S., Wood, D.A., Sadatshojaei, E. et al. Grayscale Image Statistical Attributes Effectively Distinguish the Severity of Lung Abnormalities in CT Scan Slices of COVID-19 Patients. SN COMPUT. SCI. 4, 201 (2023). https://doi.org/10.1007/s42979-022-01642-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-022-01642-8