Abstract
Lysosomal degradation of cytoplasmic components by autophagy ensures the continuous turnover of proteins and organelles and aids cellular survival during nutrient deprivation and other stress conditions. Lysosomal targeting of cytoplasmic proteins and organelles requires the concerted action of several proteins and multisubunit complexes. The core components of this machinery are conserved from yeast to humans and many of them are well-characterized; however, novel molecular players have been recently discovered and are waiting for detailed analysis. The osteopetrosis-linked PLEKHM1 protein is a lysosomal adaptor involved in autophagosome and endosome to lysosome fusion events and its role in lysosomal positioning in osteoclasts was reported together with its proposed binding partner, the relatively uncharacterized DEF8 protein. Here, we report the generation and subsequent analysis of novel mutant alleles of Drosophila plekhm1 and def8. Interestingly, the CRISPR-generated null mutations of these genes do not have any obvious effects on autophagy in Drosophila tissues, even though RNAi knockdown of these genes seems to perturb autophagy. Although these results are quite surprising and raise the possibility of compensatory changes in the case of null mutants, the new alleles will be valuable tools in future studies to understand the cellular functions of Drosophila Plekhm1 and Def8 proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autophagy is a conserved self-degradative process of eukaryotic cells by which obsolete organelles or long-lived proteins are sequestered from the cytoplasm and degraded in lysosomes (Klionsky and Codogno 2013). Autophagy is considered to occur in all cells at a basal rate; however, rapid induction of autophagy can be triggered by starvation and other factors of cellular stress. Three main types of autophagy (macro-, micro- and chaperone-mediated autophagy) have been described based on the mechanisms of lysosomal targeting of cargo destined for degradation. During the first steps of macroautophagy (hereafter referred to as autophagy) a portion of the cytoplasm is surrounded by a cup-shaped double-membrane cistern (the isolation membrane or phagophore) that ultimately seals around the sequestered material and forms a double-membrane vesicle called autophagosome. Autophagosomes then fuse with lysosomes to give rise to autolysosomes where degradation takes place. A set of evolutionarily conserved genes responsible for autophagosome formation were identified in yeast almost thirty years ago and named as Atg (autophagy-related) genes/proteins (Tsukada and Ohsumi 1993). Besides Atg proteins, many other molecular players have been shown to be necessary for processes related to autophagosomal clearance. Factors involved in these processes are also largely conserved and represent a versatile group of proteins including Rab small GTPases, the HOPS (homotypic fusion and protein sorting) tethering complex and the fusion-executing SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein receptor) proteins (Lőrincz and Juhász 2020).
The discovery of molecular players of the autophagy pathway is still in progress as novel proteins with functions in vesicular tethering have been described in the past few years. Such proteins include PLEKHM1 (pleckstrin homology domain containing protein family member 1) and DEF8 (differentially expressed in FDCP 8), the lysosome-related function of which are reported in recent studies. The function of mammalian PLEKHM1 as a lysosomal adaptor is supported by several studies and it was shown that PLEKHM1 binds to Rab7, the HOPS complex and GABARAP (Gamma-aminobutyric acid receptor-associated) family proteins, respectively (Tabata et al 2010; McEwan et al 2015; Fujiwara et al 2016; Rogov et al 2017). Through these interactions, PLEKHM1 promotes the fusion of lysosomes with late endosomes and autophagosomes. In humans and rats, mutations of PLEKHM1 lead to osteopetrosis, a disease characterized by impaired osteoclast function (Van Wesenbeeck et al 2007). Mutant osteoclasts show abrogated peripheral distribution of lysosomes indicating the role of PLEKHM1 in lysosome positioning and secretion (Fujiwara et al 2016). Importantly, this study postulates the existence of a PLEKHM1-containing protein complex that also includes DEF8, a largely uncharacterized protein.
Using the fruit fly Drosophila melanogaster as a model we reported earlier that knockdown of Plekhm1 abrogates the fusion of lysosomes with secretory granules in larval salivary glands and we also showed that Drosophila Plekhm1 directly interacts with Rab7 in yeast two-hybrid experiments (Csizmadia et al 2018; Lőrincz et al 2017). These results indicate a conserved role of Plekhm1 in lysosomal fusion events; however, human PLEKHM1 and Drosophila Plekhm1 proteins show significant differences. Fly Plekhm1 is 326 amino acids shorter as the middle part of the protein (containing several predicted domains) is missing from it when compared to human PLEKHM1. Accordingly, overall sequence identity of the proteins is only 21.8%. Exact molecular functions and the presumable involvement of Drosophila Plekhm1 in autophagy have not been investigated in detail.
In this study, we describe the generation of novel mutant alleles of the Drosophila plekhm1 and def8 loci, and assess the effect of these mutations on autophagy using fruit fly tissues. Surprisingly, no major alterations in the number and size of autophagic structures and no significant changes in the levels of autophagic cargo proteins could be detected in the generated mutants. Although these basic tests did not reproduce our observations based on RNAi knockdowns that do affect autophagy, our novel mutant alleles can be used in future studies to analyze the cellular functions of Drosophila Plekhm1 and Def8 proteins.
Results
Knockdown of Plekhm1 and Def8 leads to altered distribution of autophagic structures in fat cells
The fruit fly is an excellent model animal to analyze the functions of genes with predicted autophagic functions as developmentally programmed and stress-induced autophagic responses predictably occur in fly tissues (Nagy et al 2015). A good and common choice to investigate gene functions related to autophagy is the larval fat body with its large and easily accessible cells and with various genetic and experimental tools to visualize autophagic structures within these cells. In the fat body of well-fed third instar larvae autophagy takes place only at a basal level; however, starvation induces massive autophagic response in the tissue. Autophagosomes and autolysosomes are formed in large numbers and can be detected using fluorescent reporters of Atg8a, a widely used autophagy marker.
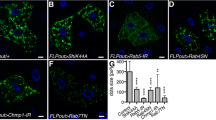
To assess the putative autophagic functions of Plekhm1 and Def8 first we applied an RNA-interference-based mosaic approach in the larval fat body (Fig. 1). The combination of the heat-shock-driven Flip recombinase with a special genetic cassette leads to the generation of fat body cells expressing RNAi-constructs targeting particular genes together with GFP as a cell marker. Autophagic structures in the whole tissue are labeled by the 3xmCherry-Atg8a reporter and their pattern can be easily compared between GFP-marked knockdown cells and the neighboring GFP-negative control cells. Using this simple and robust approach we were able to detect a pronounced effect of Plekhm1 and Def8 knockdown on the pattern of autophagic structures in fat cells of starved larvae. In control cells, smaller autophagosomes and larger, brightly fluorescent autolysosomes can be detected scattered throughout the cytoplasm. In contrast, expression of RNAi constructs targeting Plekhm1 (Fig. 1a) or Def8 (Fig. 1b) leads to the perinuclear arrangement of autophagic structures. Importantly, elongated 3xmCherry-Atg8a-positive entities probably representing clusters of autophagosomes (arrows in Fig. 1a and b) can also be detected in knockdown cells in both cases. However, statistical analysis (not shown here) did not reveal significant differences between control and knockdown cells neither in the number of autophagic structures nor in the area covered by them. Nevertheless, the detected rearrangements in the pattern of autophagic structures point to the involvement of Drosophila Plekhm1 and Def8 proteins in autophagy in larval fat cells, in line with data from mammals (McEwan et al 2015; Fujiwara et al 2016).
Knockdown of Plekhm1 and Def8 leads to altered distribution of autophagic structures in fat cells. Autophagic structures appear in fat cells due to starvation and are labeled by the autophagy reporter 3xmCherry-Atg8a. GFP-marked fat cell clones express the indicated knockdown constructs targeting Plekhm1 a or Def8 b. In GFP-expressing cells (outlined in panels a’ and b’) autophagic structures (presumably autophagosomes) seem to be perinuclearly arranged in contrast to the surrounding control cells. Arrows in a’ and b’ indicate elongated 3xmCherry-Atg8a-positive structures that most probably represent clusters of autophagosomes. Scale bar: 20 µm
The novel plekhm1 d14 and def8 d43 mutations represent null alleles
To obtain stronger evidence for the functions of Plekhm1 and Def8 in autophagy, we wanted to analyze the mutant phenotypes of both genes. As no mutant alleles have been reported so far, we set out to disrupt plekhm1 and def8 using the CRISPR/Cas9 system. We used a double guide RNA approach (Kondo and Ueda 2013) to facilitate efficient generation of gene deletions: gRNAs were designed to target the 5’ end and middle of the coding sequences of plekhm1 and def8.
During the screening of more than one hundred established candidate strains in the case of both genes no lethal mutations were isolated. However, PCR-genotyping identified several independent viable deletion mutants of plekhm1 and def8. Two examples of PCR fragments with smaller size than the wild-type cases are illustrated in Fig. 2a and b. Sequence analyses showed that approximately the first halves of the genomic regions of the targeted genes are deleted in the case of the depicted alleles, as expected based on the double gRNA targeting (Fig. 2c). Mutations both with and without frameshifts were identified (Fig. 2d) in both cases: in the case of the plekhm1d18 and def8d120 alleles in-frame deletions within the coding sequences produce proteins lacking the 16–431 and the 12–305 amino acids, respectively, while in the case of the plekhm1d14 and def8d43 alleles deletions lead to frameshifts that introduce scrambled sections and premature stop codons after amino acids 11 and 12, respectively. Such aberrant transcripts may also be eliminated by nonsense-mediated decay, and truncated proteins haphazardly translated from the remaining coding sequence are not likely to retain functions of the whole protein. Thus, we consider the plekhm1d14 and def8d43 mutations as null alleles of plekhm1 and def8 and we focused on these two alleles in our next experiments. Animals homozygous for these alleles are viable, the adults normally emerge from the pupal cases, they are fertile and no obvious external phenotypic alterations can be visually identified under the stereomicroscope.
The novel plekhm1d14 and def8d43 mutations represent null alleles. a and b Results of PCR carried out using genomic DNA isolated from heterozygous plekhm1 and def8 mutant animals. c Schematic views of the genomic regions containing the plekhm1 and def8 loci. Brackets show the deleted segments of the genes in the case of the indicated alleles. d ClustalW analysis of wild type versus mutant Plekhm1 and Def8 protein sequences. Due to in-frame mutations in case of the plekhm1d18 and def8d121 alleles, wild-type amino acid sequences (highlighted in green) are preserved after a deletion in the DNA-sequences of both genes. In case of the plekhm1d14 and def8d43 alleles, nonsense mutations introduce premature stop codons (indicated with red asterisks) into the protein sequences after a short, scrambled protein section (underlined with red)
The novel plekhm1 and def8 null alleles show no obvious autophagy defects
We again first turned to mosaic analysis as it allows one to compare mutant and control cells side-by-side within the same tissue, thus eliminating the effect of phenotypic variance between animals with the same genotype (Xu and Rubin 1993). To enable such analyses in the case of our novel loss-of-function alleles of plekhm1 and def8 we made use of the mitotic recombination system.
After recombining the plekhm1d14 and def8d43 alleles with the appropriate FRT-containing chromosomes for Flp-mediated generation of homozygous null mutant cells, we set up genetic crosses to pursue mosaic analyses of the generated alleles (Fig. 3a–c). In our experimental setup, homozygous plekhm1d14 or def8d43 mutant fat body cells lack GFP-expression and they are surrounded by GFP-expressing heterozygous mutant (control) cells. To our surprise, the size and distribution of autophagic structures were largely unaffected in homozygous plekhm1d14 fat cell clones of starved larvae. No apparent differences could be detected either in the pattern of the mCherry-Atg8a reporter (Fig. 3a) or in the case of the acidophilic dye Lysotracker Red (Fig. 3b). Similarly, homozygous def8d43 mutant fat cell clones of starved larvae also displayed normal mCherry-Atg8a pattern (Fig. 3c). Thus, mutant plekhm1d18 and def8d43 fat body cells failed to reproduce the phenomena observed in the case of the knockdown experiments. Interestingly, a pronounced increase in cell size of homozygous plekhm1d14 clones was apparent (Fig. 3a, b), although we did not detect such effect either in Plekhm1- or Def8-knockdown (Fig. 1a, b), or in def8d43 mutant cells (Fig. 3c).
The novel plekhm1 and def8 null alleles show no obvious autophagy defects. a–c Mosaic analysis in larval fat tissue. Autophagic structures appear in fat cells due to starvation and are labeled by the autophagy reporter mCherry-Atg8a in a and c or the acidophilic vital dye Lysotracker Red in b. Cells marked by the absence of GFP are homozygous for the indicated alleles of plekhm1 in a and b or def8 in c, respectively. The pattern of autophagic structures shows virtually no difference between GFP-minus mutant and GFP-expressing control cells. A pronounced cell-size increase can be observed in the case of plekhm1-mutant, but not in the case of def8-mutant cells compared to their neighboring control cells [mutant and control cells are outlined with white and yellow dashed lines, respectively, in panels a’, b’ and c’]. Scale bar: 20 µm. d Western blot analysis of protein lysates isolated from 3-days-old adult flies. While the atg55cc5 mutant (used as a positive control) shows marked accumulation of unlipidated Atg8a (Atg8-I) and Ref(2)P compared to the wild-type control, no increased levels of these proteins can be observed in age-matched plekhm1 or def8 mutant adult flies
Assessing the amount of Atg8a and Ref(2)P proteins is a common test to monitor the integrity of the autophagy pathway in fruit fly tissues (Pircs et al 2012). Lipidated Atg8a (ortholog of mammalian LC3 and GABARAP family of proteins) is anchored to both sides of the growing phagophore membrane and half of the protein pool is subsequently degraded in autolysosomes. The Ref(2)P (p62 or SQSTM1 in mammals) protein is also continuously degraded during autophagy as it recruits ubiquitylated cargo to Atg8a bound to the phagophore through its LIR (LC3-interacting region) motif. Thus, accumulation of Ref(2)P and Atg8a are hallmarks of defective autophagy. Using protein lysates of 3-days-old adult flies we carried out western blots to assess the amount of Atg8a and Ref(2)P in the novel plekhm1 and def8 mutants (Fig. 3d). In the wild-type case, relatively small amount of the Ref(2)P protein can be detected, and Atg8a-I (cytoplasmic, non-lipidated Atg8a) and Atg8a-II (membrane-bound, lipidated Atg8a) are present in equal amounts in the sample. In the autophagy-defective atg55cc5 mutant (used as positive control) conjugation of Atg8a to the phagophore membrane is disturbed, and as a consequence, non-lipidated Atg8a-I accumulates, and Ref(2)P also shows marked accumulation. In contrast, we could not detect increased levels of Atg8a or Ref(2)P proteins in the absence of Plekhm1 or Def8. Taken together, these results show that autophagy is not impaired in plekhm1 and def8 mutant animals.
Discussion
In this study, we aimed to investigate the functions of Drosophila Plekhm1 and Def8 in the context of autophagy using independent loss-of-function approaches. The basis of this work was our initial observation of altered autophagy in larval fat body cells expressing knockdown constructs for Plekhm1 and Def8. These results showed the perinuclear clustering of autophagic structures in both cases. Altered position of autophagic vesicles could reflect on a role of Plekhm1 and Def8 in vesicular positioning.
Based on these promising results we went on to create mutant alleles of plekhm1 and def8, and we successfully generated several deletion (null) alleles for both genes. Surprisingly, our novel mutants did not show any detectable autophagy defects and failed to reproduce the phenomena observed in our knockdown experiments.
One possible explanation of this controversy is that the observed changes in the knockdown cases might be artifacts originating from off-target effects of the used RNAi-constructs. Reinforcing these results with the use of additional RNAi-constructs together with the quantitative analysis of their knockdown efficiencies could argue against this possibility. However, the similar alterations in the case of two different knockdown constructs that target different proteins with presumably similar functions make this possibility very unlikely anyway. Accordingly, in silico off-target search using the online GESS tool (flyrnai.org/gess) identified no additionally targeted genes with potential roles in autophagy or other vesicular trafficking processes. Thus, we assume that the knockdown phenotypes reflect on autophagy-related roles of Plekhm1 and Def8.
Then, why there are no detectable autophagy defects in the case of our novel mutants? A possible explanation of this is that N-terminally truncated forms of Plekhm1 and Def8 (transcribed from an alternative transcription start site in the mutant loci) might be present in mutant animals, and that these short proteins may retain some residual functions. As we did not address this question experimentally, we cannot exclude this possibility; however, such scenario seems not to be likely. Mammalian PLEKHM1 is an adaptor protein with functional domains at both ends and Drosophila Plekhm1 is also predicted to contain these domains. As these domains are required to form a molecular bridge between two vesicles, a truncated form of Plekhm1 lacking more than a half of the wild-type protein would not be able to fulfill its function. Def8 also contains modular domains at both ends, the absence of which likely leads to the loss of the molecular functions of the protein. Thus, we believe that our generated alleles are likely null alleles that impair the functions of plekhm1 and def8.
Is it possible that the overall cellular effect of the RNAi-mediated knockdown of a particular gene is stronger than the effect of the loss-of-function mutation of the same gene? There is a growing body of evidence that such phenomenon may be the consequence of compensatory responses induced by the loss of a gene, but not in the case of its post-transcriptional targeting (Kok et al 2015; Rossi et al 2015). These mechanisms contribute to the survival of the organism in the changing environment by alleviating the effects of genetic disturbance and are summarized under the term of genetic compensation (El-Brolosy and Stainier 2017; Peng 2019). Compared to mammals, the Drosophila genome shows a smaller extent of genetic redundancy in general; however, it is not devoid of genes with similar sequences and redundant functions. In the case of plekhm1, prd1 was identified as the ortholog of mammalian PLEKHM2. However, sequence comparison shows that only a relatively low level of identity (35%) can be detected in the coding sequences of Drosophila plekhm1 and plekhm2. Accordingly, only 9% of identity was found in the protein sequences of the genes, and the predicted domain structures of the proteins also differ largely. Thus, Drosophila Plekhm2 may not compensate for the loss of Plekhm1.
Interestingly, FlyBase reports that Plekhm1, Def8 and Rubicon are paralogous genes. Indeed, amino acid sequence of Def8 shares 33.7% similarity with Plekhm1 and 29.4% similarity with Rubicon. Domain structures predicted by InterPro are also quite similar, especially in the C-terminal region where all three proteins contain a putative zinc-RING and/or ribbon domain. Such similarities raise the possibility that Plekhm1, Def8 and Rubicon might have redundant functions. Thus, effects of a loss-of-function mutation affecting one of the three genes might be at least partially compensated by the other two functional loci. Importantly, FlyBase reports that Drosophila Rubicon is predicted to be involved in the negative regulation of autophagy and is capable of phosphatidylinositol phosphate binding that enables membrane recruitment. Thus, Rubicon could be a potential candidate through which genetic compensation might act. This hypothesis could be tested by generating double or triple mutants; however, this exceeds the scope of the present study. Analyzing the possible functional redundancy between Drosophila Plekhm1, Def8 and Rubicon could be a logical and interesting sequel of our work presented here.
Although we cannot give a clear explanation for the absence of autophagy defects in the case of the newly generated plekhm1 and def8 alleles, we believe that these novel alleles will be valuable genetic tools to investigate the functions of plekhm1 and def8, two genes with uncharacterized roles in Drosophila tissues.
Materials and methods
Drosophila work
Flies were kept on standard yeast/cornmeal/agar media at 25 °C and a 12-h light/12-h dark daily cycle, under uncrowded conditions. The following Drosophila stocks were used: Plekhm1 RNAi (VDRC42065), Def8 RNAi (BDSC28312), nos-phiC31; attP40 (BDSC25709), nos-phiC31; attP2 (BDSC25710), Act-Cas9 (BDSC54590), plekhm1 Df (BDSC7558), def8 Df (BDSC7558), FRT42D (BDSC1802), FRT80B (BDSC1988), plekhm1d14, plekhm1d18, def8d43, def8d121, w1118 (BDSC3605), atg55cc5 (Kim et al 2016) and stocks for clone generation: {hs-Flp; 3xCherry-Atg8a, UAS-GFP; Act-CD2-Gal4}, {hs-Flp; cg-Gal4, FRT42D, UAS-GFPnls}, {hs-Flp; cg-Gal4, UAS-mCherryAtg8a, FRT42D, UAS-GFPnls}, {hs-Flp; cg-Gal4, UAS-mCherry-Atg8a, FRT80B, UAS-2xEGFP}. Recombination of the generated mutant alleles to FRT-containing chromosomes was carried out by standard genetic crosses. To generate mutant fat body clones FRT42D, plekhm1d14 and FRT80B, def8d43 flies were crossed to the appropriate clone-generating stocks and F1 embryos were heat-shocked 2–4 h after egg laying for 1 h at 38 °C to induce mitotic recombination. In knockdown analyses, RNAi cells were generated spontaneously in larvae carrying hs-Flp, UAS-Dcr2, Actin > CD2 > Gal4, 3xCherry-Atg8a and the respective UAS-RNAi construct. To enhance RNAi efficiency, progeny were kept at 29 °C. To induce autophagy with amino acid deprivation, well-fed third instar larvae were placed in 20% sucrose solution in a microfuge tube for 4 h.
Generation of novel plekhm1 and def8 alleles
We applied the double gRNA approach reported to be highly effective in Drosophila (Kondo and Ueda 2013). PAM-sequences within the plekhm1 and def8 loci were identified using the Cas9 target finder (https://shigen.nig.ac.jp/fly/nigfly/cas9/cas9TargetFinder.jsp), gRNA-s were designed to target the 5’ end and middle of the coding sequences. The following primers were used - Plekhm1-39-F: CTTCGAAACGGGAGAATGTCGTGA; Plekhm1-39-R: AAACTCACGACATTCTCCCGTTTC; Plekhm1-591-F: CTTCGGAGATCTGGGAGAAGTTTC; Plekhm1-591-R: AAACGAAACTTCTCCCAGATCTCC; Def8-15-F: CTTCGGACAGTCTCACGAGCATTC;Def8-15-R: AAACGAATGCTCGTGAGACTGTCC; Def8-119-F: CTTCGCCACGCAGAGTGTCGCGCA; Def8-119-R: AAACTGCGCGACACTCTGCGTGGC. Annealed oligos were cloned into the attB-containing pBFv-U6.2B vector and the created plasmids were injected into embryos with landing sites on the 3rd (attP2) or 2nd chromosome (attP40) in the case of Plekhm1 and Def8 gRNA plasmids, respectively. Stable transgenic lines were established and gRNA-expressing males were crossed to Act-Cas9 females. F2 males were crossed to females carrying a deficiency overlapping with plekhm1 or def8; no lethal candidates were isolated. Genomic DNA was extracted from F3 progeny and PCR-genotyping was carried out to screen for deletions using the following primers: Plekhm1-del-F: GACCCTCGGAAAACCACAAC; Plekhm1-del-R: GTACTCCCTGCCAGCAAACT; Def8-del-F: CAATTGCTCGCATCCCTGG; Def8-del-R: GTACTCCCTGCCAGCAAACT. DNA and protein sequence analyses were carried out using SnapGene Viewer and ClustalW, respectively.
Microscopy
Lysotracker Red staining of larval fat bodies isolated from third instar larvae was carried out as previously described (Szatmári et al 2014). Images were taken with a Zeiss AxioImager.M2 fluorescent microscope equipped with ApoTome.2 and Hamamatsu ORCAFlash 4.0 LT digital camera using an 0.95 NA 40 × Plan-Apochromat objective. Images were captured and their brightness and contrast were adjusted with the Zeiss ZEN Pro software.
Western blot
Western blots were carried out as described (Takáts et al 2013), using protein samples of 3-days-old adults. The following antibodies were used in a 1:2000 dilution: rabbit anti-Atg8a (abcam #109364), mouse anti-Tubulin (DSHB #AA4.3), rabbit a-Ref(2)P (Pircs et al 2012).
References
Csizmadia T, Lőrincz P, Hegedűs K, Széplaki S, Lőw P, Juhász G (2018) Molecular mechanisms of developmentally programmed crinophagy in Drosophila. J Cell Biol 217(1):361–374. https://doi.org/10.1083/jcb.201702145
El-Brolosy MA, Stainier DY (2017) Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet 13(7):e1006780. https://doi.org/10.1371/journal.pgen.1006780
Fujiwara T, Ye S, Castro-Gomes T, Winchell CG, Andrews NW, Voth DE, Zhao H (2016) PLEKHM1/DEF8/RAB7 complex regulates lysosome positioning and bone homeostasis. JCI Insight. https://doi.org/10.1172/jci.insight.86330
Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, Burmeister M (2016) Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife 5:e12245. https://doi.org/10.7554/eLife.12245
Klionsky DJ, Codogno P (2013) The mechanism and physiological function of macroautophagy. J Innate Immun 5(5):427–433. https://doi.org/10.1159/000351979
Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Lawson ND (2015) Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 32(1):97–108. https://doi.org/10.1016/j.devcel.2014.11.018
Kondo S, Ueda R (2013) Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195(3):715–721. https://doi.org/10.1534/genetics.113.156737
Lőrincz P, Juhász G (2020) Autophagosome-lysosome fusion. J Mol Biol 432(8):2462–2482. https://doi.org/10.1016/j.jmb.2019.10.028
Lőrincz P, Tóth S, Benkő P, Lakatos Z, Boda A, Glatz G, Juhász G (2017) Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol 216(7):1937–1947. https://doi.org/10.1083/jcb.201611027
McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Dikic I (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57(1):39–54. https://doi.org/10.1016/j.molcel.2014.11.006
Nagy P, Varga Á, Kovács AL, Takáts S, Juhász G (2015) How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods 75:151–161. https://doi.org/10.1016/j.ymeth.2014.11.016
Peng J (2019) Gene redundancy and gene compensation: an updated view. J Genet Genom 46(7):329–333. https://doi.org/10.1016/j.jgg.2019.07.001
Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G (2012) Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS ONE 7(8):e44214. https://doi.org/10.1371/journal.pone.0044214
Rogov VV, Stolz A, Ravichandran AC, Rios-Szwed DO, Suzuki H, Kniss A, Löhr F et al (2017) Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep 18(8):1382–1396
Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524(7564):230–233. https://doi.org/10.1038/nature14580
Szatmári Z, Kis V, Lippai M, Hegedűs K, Faragó T, Lőrincz P, Sass M (2014) Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Mol Biol Cell 25(4):522–531. https://doi.org/10.1091/mbc.e13-10-0574
Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, Yoshimori T (2010) Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell 21(23):4162–4172. https://doi.org/10.1091/mbc.e10-06-0495
Takáts S, Nagy P, Varga Á, Pircs K, Kárpáti M, Varga K, Juhász G (2013) Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol 201(4):531–539. https://doi.org/10.1083/jcb.201211160
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333(1–2):169–174. https://doi.org/10.1016/0014-5793(93)80398-E
Van Wesenbeeck L, Odgren PR, Coxon FP, Frattini A, Moens P, Perdu B, Van Hul W (2007) Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Investig 117(4):919–930. https://doi.org/10.1172/JCI30328
Xu T, Rubin GM (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117(4):1223–1237. https://doi.org/10.1242/dev.117.4.1223
Funding
Open access funding provided by ELKH Biological Research Center. This work was supported by the National Research Development and Innovation Office (NKFIH) of Hungary with PD135587 grant to TM and GINOP-2.3.2-15-2016-00032, Elvonal KKP129797 and National Laboratory for Biotechnology program grants to GJ.
Author information
Authors and Affiliations
Contributions
TM, EL, DF and PL carried out the experiments. TM, EL and GJ wrote the manuscript. GJ oversaw the research.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Consent to participate
All authors gave their consent to participate in the work presented in the current manuscript.
Consent for publication
All authors gave their consent for the publication of the manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maruzs, T., Lakatos, E., Feil-Börcsök, D. et al. Isolation and characterization of novel plekhm1 and def8 mutant alleles in Drosophila. BIOLOGIA FUTURA 73, 149–155 (2022). https://doi.org/10.1007/s42977-022-00118-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-022-00118-3