Abstract
Lysosomal-dependent self-degradative (autophagic) mechanisms are essential for the maintenance of normal homeostasis in all eukaryotic cells. Several types of such self-degradative and recycling pathways have been identified, based on how the cellular self material can incorporate into the lysosomal lumen. Ubiquitination, a well-known and frequently occurred posttranslational modification has essential role in all cell biological processes, thus in autophagy too. The second most common type of polyubiquitin chain is the K63-linked polyubiquitin, which strongly connects to some self-degradative mechanisms in the cells. In this review, we discuss the role of this type of polyubiquitin pattern in numerous autophagic processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cellular self-digestive mechanisms (autophagy) have a main contribution in the fast-cellular response to the changeable environment and maintain the homeostatic equilibrium of the eukaryotic cells. The essential function of autophagic mechanisms is to degrade several types of cellular components such as cell organelles, signalling molecules and well-defined parts of the cytoplasm. Moreover, self-digestive pathways have another importance in the cellular life: eukaryotic cells use autophagic mechanisms to recycle the molecules of the degraded self-material into the cytoplasm. The unnecessary or injured components of the cells can be degraded by the endo-lysosomal (lysosome dependent) system (autophagy) and the Ubiquitin–Proteasome (lysosome independent) System—UPS (Bhattacharyya et al. 2014; Kwon and Ciechanover 2017; Marzella et al. 1981; Weckman et al. 2014). There are four main types of autophagic mechanisms based on how the cytoplasmic material incorporates into the lysosomal lumen: macroautophagy, microautophagy, chaperon-mediated-autophagy and crinophagy. The lysosome dependent autophagic mechanisms are responsible for the breakdown of the significant cytoplasmic material (macromolecules) and cell organelles such as peroxisomes, endoplasmic reticulum (ER), part of the nucleus, secretory granules, mitochondria and damaged lysosomes (Csizmadia and Juhász 2020; Galluzzi et al. 2017; Marzella et al. 1981; Weckman et al. 2014). In contrast the Ubiquitin–Proteasome System is only capable of removing and recycling short half-life proteins from the cytoplasm, which control several signalling pathways (Bhattacharyya et al. 2014; Kudriaeva and Belogurov 2019; Kwon and Ciechanover 2017). These self-degradative mechanisms have high biomedical significances because various types of disorders are connected to the reduced function of autophagy and UPS such as diabetes, fatty liver disease (FLD), cancer, accelerated aging, neurodegenerative, infectious, and vascular diseases (Bhattacharjee et al. 2019; Coux et al. 2020; Ruocco et al. 2016). Interestingly, autophagy and UPS work together and complement each other for the effective degradation of cellular components which leads to normal homeostatic equilibrium during various internal or external effects (Dikic 2017; Kwon and Ciechanover 2017; Lőw et al. 2013). Importantly, ubiquitin, as a frequent posttranslational modifying molecule, also has a role in proteasome independent mechanisms such as epigenetic regulation, DNA damage response, cell cycle control, diverse signalling pathways, intracellular vesicular trafficking and autophagic mechanisms. The differential role of ubiquitin code in these cell biological processes is connected to the variety of the ubiquitin pattern on the target proteins (Clague and Urbé, 2017; Swatek and Komander 2016). In this review we discuss the role of the second most common type of polyubiquitin, the Lysine 63 (K63) connected polyubiquitin in several autophagic mechanisms.
Ubiquitin and the ubiquitination system
Ubiquitination is one of the most important and reversible covalent posttranslational modification of proteins. The name “ubiquitin” was coined by Gideon Goldstein, the discoverer of this modifying protein, reflecting that ubiquitination or similar mechanisms could be observed in almost any organism (Goldstein et al. 1975). During this process, an isopeptide bond is formed between a Lysine (K) residue of the target protein and the C-terminal glycine of ubiquitin, a 76 amino acid long, evolutionarily conserved protein (Fig. 1). The isopeptide contact is an amide bond, that is chemically similar to the classical peptide bond, which is created by a condensation reaction between the α-amino- and the C1 carboxyl group of two amino acids (Nelson and Cox 2017a). In the isopeptide bond however, at least one of the reacting amino acids participates with the amino group on its side chain, instead of the α-amino group (Alberts et al. 2015; Nelson and Cox 2017c). The formation of these peptide bonds is thermodynamically unfavourable, so it does not happen spontaneously in the cellular environment. On the other hand, through the covalent modification (activation) of a carboxyl group utilising the free energy change in ATP hydrolysis, the process could be made energetically favourable (Nelson and Cox 2017b). Importantly, ubiquitin molecules can also connect with each other between specific Methionine (Met) and Lysine (K) residues: Met1, K6, K11, K27, K29, K33, K48 and K63 via isopeptide bond (Csizmadia and Lőw 2020).
Structure of the ubiquitin molecule (Zheng and Shabek 2017). The Lysine residues responsible for the formation of polyubiquitin chains are marked by green, the first Methionine residue is marked by olive green, and the C-terminal Glycine is marked by yellow. It is visible, that the C-terminal carboxyl group and the M1 and K63 residues are on the opposite sides of the protein, which makes the M1- and K63-linked polyubiquitin chains elongated and flexible. The most abundant polyubiquitin chain type is composed of ubiquitin moieties linked by the K48-residue (located closer to G76) are more compacted and rigid than M1- and K63-linked polymers. (PDB ID: 1UBQ. Modified in PyMol by the authors)
Ubiquitination is a process that involves three main enzymes: an E1 ubiquitin-activating, an E2 ubiquitin-conjugating, and an E3 ubiquitin-ligase enzyme. In the first step, using an ATP molecule, an ubiquitin-adenylate intermediate is formed, that activates the C-terminus of the ubiquitin. Then a thioester bond is formed between the E1 enzyme and the activated intermediate, thus the C-terminus of the ubiquitin molecule will be covalently bound to the E1 (Pickart 2001). Next, ubiquitin is transferred to the E2 enzyme, with another thioester bond forming. All of the E2 enzymes have ubiquitin-conjugating catalytic domain (UBC) that contain the indispensable serine residue in the active site (van Wijk and Timmers 2010). Finally, the recognition and recruitment of the target protein is done by the E3 enzyme (Fig. 2). The group of E3 enzymes consist of several protein families that serve the same purpose through different mechanisms:
-
1. The Really Interesting New Gene (RING) type E3 enzymes are not attached covalently to ubiquitin during ubiquitination, but by forming a complex with the ubiquitin bound E2 and the substrate. They mediate the direct transfer of ubiquitin from the E2 to the target Lysine substrate (Zheng and Shabek 2017).
-
2. Homologous to the E6AP Carboxyl Terminus (HECT) type E3 enzymes themselves form a thioester bond with the activated ubiquitin before the ligation to the substrate.
-
3. Recently, a new type of E3 enzymes was described, which are called RING-between-RING (RBR) ubiquitin-ligases. This type of E3 enzymes are functionally the hybrids of RING and HECT ligases (Zheng and Shabek 2017).
The mechanism of the ubiquitination. An E1 enzyme activates ubiquitin by the free energy change in an ATP molecule and transfers it on an E2 enzyme. The ubiquitin bound E2 forms a complex with an E3 enzyme that recruits it to the substrate protein, and the ubiquitin is transferred to the substrate. The substrate molecule can be monoubiquitinated by a single ubiquitin moiety, or polyubiquitinated by a chain of ubiquitin molecules ligated to each other through one of their Lysine residues. The structure of the polyubiquitin chain depends on the linking Lysine residue. By ubiquitination on more than one residue, branched chains could be produced. Ubiquitin-Binding Proteins (UBPs) can specifically recognize (read) different ubiquitin patterns (code) and facilitate signal transduction. The removal of ubiquitin from the substrate is catalysed by deubiquitinase enzymes
Importantly, E3 enzymes sometimes have cooperating adaptor proteins, which help the recognition of specific substrates (Kong et al. 2020; Li et al. 2015). Moreover, multi-subunit E3 complexes are also known, like the well characterised Anaphase Promotion Complex or Cyclosome (APC/C) (Badarudeen et al. 2021).
Usually only one E1 enzyme is coded in the genome of most living organisms (generally named Ubiquitin Activating Enzyme 1 or UBA1), or two in vertebrates (UBA1 and UBA6) (Clague et al. 2015). In the human proteome, there are about 40 E2 enzymes, and potentially several hundreds of the E3s, that need to specifically bind to their substrates (Clague et al. 2015; Zheng and Shabek 2017). Between E2 and E3 enzymes, both highly specific and non-specific binding is known. Interestingly, there are special enzymes in both groups that can bind to only one protein of the other type, and several E2s and E3s, too, that can function with many interaction partners (Clague et al. 2015; Zheng and Shabek 2017).
The substrates can be differentially ubiquitylated. Monoubiquitination involves a single ubiquitin molecule ligated to the substrate, and multi-monoubiquitination means monoubiquitination at multiple sites on the target at the same time (Komander and Rape 2012). Interestingly, ubiquitin itself can be a target of ubiquitination, enabling the formation of polyubiquitin chains (Zheng and Shabek 2017). The topology and structure of the polyubiquitin chains are determined by the Lysine residue of the proximal moiety, to which the G76 of the distal ubiquitin gets ligated (Fig. 1).
The linkage types are thus discriminated by the proximal moiety’s residue: K6-, K11-, K27-, K29-, K33-, K48-, K63-linked polyubiquitin chains can be assembled by isopeptide bonds (Csizmadia and Lőw 2020; Swatek and Komander 2016). A classical peptide bond between the N- and C-termini of two ubiquitin moieties can also be formed, that is either called Methionine1 (Met1)-linked polyubiquitin because of the determining proximal residue, or linear ubiquitin which refers to the continuity of the polypeptide chain between moieties (Rieser et al. 2013). There are hydrophobic patches and protein interaction sites on the surface of ubiquitin, which either take part in shaping the polyubiquitin chain, or facilitate interaction with other proteins (Komander and Rape 2012). Polyubiquitin chains linked by the same Lysine residues are called homotypic chains, and chains that contain different linkage types are called heterotypic chains (Csizmadia and Lőw 2020; Komander and Rape 2012; Swatek and Komander 2016). More than one polyubiquitin chain could be attached to the substrate proteins, that also applies to ubiquitin itself: by ligating ubiquitin to more than one capable residue of the proximal moiety, branched chains could be created (Swatek and Komander 2016). Posttranslational modifications, like phosphorylation and SUMOylation can also affect ubiquitin, too. All of these taken into consideration, it is evident that using ubiquitin as a signal, a great variety of different patterns (codes) could be created on target proteins, that enables diverse and sophisticated signalling through this process. The Ubiquitin-binding Proteins (UBPs) are able to differentially bind to polyubiquitin chains, and so the information coded into the ubiquitin signal could be carried on to other components of a signalling pathway. By 2019, 29 different Ubiquitin-binding domains (UBDs) have been discovered, and this number will probably increase even further in the future (Radley et al. 2019).
Moreover, some early publication mentions the equivalent role of the E4 enzymes in the assembly of polyubiquitin chain on the target proteins. Interestingly, in the yeast, UFD2 E4 enzyme has function in cell stress tolerance, but it is not essential in cell viability (Hoppe 2005; Koegl et al. 1999).
Proteases that can bind to ubiquitin and cleave the isopeptide bond between two moieties or the substrate and the ubiquitin connected to it, are called deubiquitinases (DUBs) (Komander et al. 2009a). Most DUBs are specific for hydrolysing isopeptide bonds, as those are chemically different from peptide bonds. According to their catalytic domain, DUBs can be classified into five families: the Ubiquitin Specific Proteases (USP), Ubiquitin Carboxyl-terminal hydrolases (UCH), ovarian tumour proteases (OTU), and Josephine family DUBs are Cisteine-proteases, while JAB1/MPN/MOV34 (JAMM) family proteases are Zn2+ metalloproteases (Clague et al. 2013; Komander et al. 2009a). Some DUBs are specific for one or few polyubiquitin linkage types, but non-specific DUBs also exist. DUBs can specialize in cleaving the distal ubiquitin moiety, a bond inside the chain, or the bond between the substrate and the proximal ubiquitin.
The structure and role of K63-linked polyubiquitin
The second most abundant polyubiquitin chain type in cells, with primarily non-degradative roles, is linked through the K63 residue. Lysine 63 is located close to the M1 residue, opposite to G76. Linkage through K63 residues gives a flexible and elongated structure to the polyubiquitin chain (Komander et al. 2009b). Because M1 and K63 are so close to each other, the polyubiquitin chains linked through them are also similar in structure. Komander et al. confirmed in 2009 that there are no hydrophobic interactions between the proximal and distal ubiquitin moieties in these linkage types, they are only connected to each other through their isopeptide bond. This leaves the hydrophobic patch around Isoleucine 44 open to interaction with other proteins, unlike K48-linked polyubiquitin, where the moieties also interact with each other through this site, resulting in a differently shaped polyubiquitin structure. They predicted that two moieties linked through K63 or M1 will give an elongated structure compared to the K48-linked diubiquitin, which they confirmed through X-ray crystallography. They also found that these chains were much more flexible than K48-linked polyubiquitin, which enables a greater variety of interactions with ubiquitin binding proteins. In the same year, it was observed that the length of the K63-linked tetraubiquitin however was shorter in length, than what was originally predicted (although still significantly longer that its K48-linked counterpart) (Datta et al. 2009). This difference is thought to be caused by the flexibility of the chain, as the hydrophobic patches remained exposed. The unique structural and chemical differences of the K63-linked polyubiquitin allow it to be recognised by different proteins, and to convey different information than other linkage types. That’s why K63-linked polyubiquitination can have proteasome-independent, non-degradative roles in the cells (Swatek and Komander 2016).
So far, only one K63-specific E2 enzyme has been identified, that is called Ubiquitin-conjugating enzyme 13 (UBC13) in yeast, and its ortholog in humans Ubiquitin-conjugating enzyme E2 N (UBE2N) (Hodge et al. 2016). UBC13 is only functional in heterodimeric form. In the nucleus, it forms a complex with MMS2 (Methyl MethaneSulfonate sensitivity 2), and in the cytoplasm, with UEV1A (Ubiquitin Conjugating Enzyme E2 V1). Both are E2-like proteins lacking the catalytic activity of an E2 enzyme, structurally similar to each other. UBC13 can work together with a range of E3 enzymes, but there is a possibility that not all E3 enzymes need UBC13 to be able to synthetise K63-linked polyubiquitin chains.
Not only the synthesis, but the cleavage of K63-linked polyubiquitin too, requires specific enzymes. Deubiquitinases usually show a degree of specificity towards some types of polyubiquitin chains (Komander et al. 2009b). Even though M1-linked polyubiquitin chains are structurally very similar to K63-linked chains, the M1-specific deubiquitinases (DUBs) usually do not catalyse the hydrolysis of the K63-linked chains’ isopeptide bonds since the linear chains’ chemical properties are different. The only known exception is Cylindromatosis (CYLD) USP-family deubiquitinase, which binds to and cleaves M1- and K63-linked chains alike. Komander et al. found that although the different DUB families have distinctive structures, they are not specific for one type of linkage in the family level. That means that K63-specific enzymes can be found across several DUB families. The USP family of DUBs are usually capable of cleaving both K48- and K63-linked polyubiquitin with a similar efficiency, OTU family enzymes are usually more specific towards one linkage type or another. The JAMM metalloprotease investigated in this research, AMSH (Associated Molecule with the SH3 domain of STAM), was specific for K63-linkages only. It is evident though that K63-specific DUBs have evolved in most of the DUB families, and this linkage can be cleaved by more than one mechanism, both by cysteine and metalloproteases (Komander et al. 2009b) (Table 1).
The diversity of ubiquitin as a signal
Ubiquitin is best-known of its degradative functions. Cytoplasmic proteins that are incorrectly folded, lose or change their functions or no longer needed in the cells, usually get degraded in the 26S proteasome. These proteins get selected for degradation by K48-linked polyubiquitination (Hershko and Ciechanover 1998). Because of this primary function, some people call ubiquitin “the kiss of death”. Like K63-linked polyubiquitination, K48-linked chains can also be found as part of signalling pathways: as some signalling molecules go through K48-linked polyubiquitin-mediated proteasomal degradation, the output of the transduction could change. A pathway could be activated if inhibitor proteins get ubiquitinated and degraded, or it could stop the transduction by targeting active enzymes for degradation, after the signal disappears. K63-linked chains instead usually modulate the function of proteins or serve as platform for subsequent signalling steps, instead of mediating degradation (Skaug et al. 2009; Zhang et al. 2017).
The structure and function of K48- and K63-linked polyubiquitin are quite well known and understood compared to other types of linkages, which are less abundant or harder to investigate (alternatively called atypical linkages). M1-linked polyubiquitin can be found together with K63-linked polyubiquitin chains in various immune signalling pathways (Emmerich et al. 2013; Rieser et al. 2013). K6-linked polyubiquitin has been shown to participate in DNA damage repair and JNK (c-Jun N-terminal kinase)-signalling, but our knowledge about it is still limited (Kulathu and Komander 2012; Wang et al. 2020b). K11-linked polyubiquitin is involved in cell cycle regulation, through the APC/C, which is capable of producing both K11-K48 branched polyubiquitin chains on cell cycle regulating proteins (Meyer and Rape 2014). K11 linkages have been associated with degradative roles similar to K48-linked chains. K27-linked ubiquitin chains have been found in signalling pathways of innate immunity (Peng et al. 2011; Wu et al. 2019; Xue et al. 2018) and in MEK (MAPK/ERK Kinase)/ERK (Extracellular signal-regulated kinase) signalling (Yin et al. 2019). All K27-, K29- and K33-linked polyubiquitin chains were found in the attenuation of TCR (T-Cell Receptor)-signalling (Kulathu and Komander 2012). K29- and K33-linked chains also appear in the regulation of AMPK (AMP-Activated Protein Kinase), and a deubiquitinase involved in Wnt signalling, TRABID (TRAF-BInding Domain-containing protein) is specific these two linkage types (Kulathu and Komander 2012). Investigation of these atypical chains is more difficult, therefore our knowledge about their potential roles is very limited (Table 2) (Fig. 3).
Comparison of K63-linked polyubiquitin chains with chains of different topology (Datta et al. 2009; Eddins et al. 2007; Komander et al. 2009b). A: K63-linked diubiquitin, B: M1-linked diubiquitin, C: K63-linked tetraubiquitin, D: K48-linked tetraubiquitin. Both K63- (A, C) and M1-linked (B) chains possess elongated, flexible structures, while K48-linked chains (D) are much more compact. It is visible, that ubiquitin moieties in K63- and M1-linked chains are only connected by the isopeptide bonds, leaving the hydrophobic patches open. (PDB IDs: 2JF5, 3AXC, 3HM3, 2O6V;
The function of K63 linked polyubiquitin during autophagic processes
Autophagy has an important role in maintaining cell homeostasis. Through various autophagic mechanisms, the components of the cell are broken down by itself in the lysosomes hence it is alternatively called cellular self-digestion. Basal level autophagy is part of the normal cellular metabolism, but upon different stimuli from the environment, like starvation, hypoxia or injury to organelles, the intensity of the process can greatly increase to remove harmful agents from the cytoplasm. In many cases, intense autophagy is a natural part of the developmental program of an organism, usually preceding cell death (Csizmadia et al. 2018; Denton and Kumar 2019; Parzych and Klionsky 2014). The terminal compartment of cellular material selected for autophagy called the lysosome, is a membrane-bound organelle containing multiple types of degradative enzymes, like proteases, nucleases, polysaccharide-, and lipid-degrading enzymes. The enzymatic activity of these proteins is turned on by low pH in the lumen of the lysosomes. During autophagy, the to-be-degraded cellular materials get into a common space with these enzymes, and under adequate conditions, they get decomposed into smaller, generally reusable elements which then exit from the lysosomal lumen through transporter proteins to be reintegrated into the cell metabolism. The autophagic cargo can be delivered into the lysosome by multiple mechanisms, which include macroautophagy, microautophagy, chaperone-mediated autophagy (CMA) and crinophagy. In macroautophagy, a flat, membrane-bound structure called the phagophore, forms around and engulfs the selected macroautophagic cargo, creating the double-membrane-bound autophagosome (Fig. 4). The innermost part of the autophagosome may contain only a part of the cytoplasm, some small particles and vesicles, or it could contain larger organelles, like a mitochondrion or parts of the endoplasmic reticulum. The autophagosome then fuses with a lysosome, creating an autolysosome, where the cellular components start to degrade (Parzych and Klionsky 2014). In microautophagy, an invagination is formed on the membrane of the lysosome and a small portion of the cytoplasm flows into this pocket. The invagination turns into small intraluminal vesicles of the lysosome and gets degraded inside (Galluzzi et al. 2017; Mijaljica et al. 2011). In chaperone-mediated autophagy, selected cytoplasmic proteins get unfolded into the lumen of the lysosome through chaperone-like membrane proteins of the lysosome (Cuervo and Wong 2014). Interestingly, crinophagy is a special form of autophagy, in which a no longer needed or damaged secretory granule directly fuses with a lysosome to form a special type of secondary lysosome (called crinosome), independently from autophagosome formation (Ahlberg et al. 1987; Csizmadia and Juhász 2020; Csizmadia et al. 2018). No connection between K63-linked polyubiquitin and either microautophagy or crinophagy has been uncovered yet, but K63-linked polyubiquitination likely affects the initiation of selective macroautophagy, plays a role in different selective macroautophagic pathways such as aggrephagy, mitophagy and xenophagy and potentially in some cases of chaperone-mediated autophagy, too (Erpapazoglou et al. 2014; Ferreira et al. 2015). During the earlier mentioned selective macroautophagic processes the K63 polyubiquitin has an important role in the mechanism of cargo designation and recognition by the degradative system. K63-linked polyubiquitin chains are recognised by several types of autophagic receptor proteins such as p62, NDP52 (Nuclear Domain 10 Protein 52), OPTN (Optineurin) and NBR1 (Neighbour of BRCA1 gene1) (Erpapazoglou et al. 2014). Importantly, there is a well-known type of autophagy mixed with heterophagy, during which the unnecessary or obsolete plasma membrane proteins, such as receptors, signalling molecules and transporters decorated with K63 poliubiquitin chains internalise into early endosomal membrane and sort to the lumen of multivesicular bodies and lysosomes for the fast degradation of this proteins (Erpapazoglou et al. 2014). Interestingly, the discovered K63 ubiquitination events mostly activate or enhance autophagy, in contrast to K48-linked polyubiquitination, which usually causes the proteasomal degradation of signalling molecules, therefore inhibiting autophagy.
The role of K63-linked polyubiquitin in macroautophagy. This type of polyubiquitin is suitable for the designation of the cytoplasmic components which are recognised by the autophagosomal receptors such as p62, NBR1, NDP52 and optineurin, which are the prerequisite the selective packaging of the cytoplasmic components to the autophagosomal lumen via the connection with the ubiquitin like protein Atg8. After the formation of the autophagosomes, this double membrane bounded vesicles fuse with lysosomes for the fast degradation of the assigned cytoplasmatic material
Various pathways in selective macroautophagy
The initiation phase of the autophagosome formation starts at the Unc-51 Like Autophagy Activating Kinase 1 (ULK1) complex. Under normal circumstances the mammalian Target of Rapamycin Complex 1 (MTORC1) is bound to the ULK1 complex, which keeps it inactive by phosphorylation. Upon starvation, the MTORC1 dissociates from the complex, and autophagosome formation is induced (Parzych and Klionsky 2014). The namesake subunit of the complex, the ULK1 protein can be polyubiquitinated by K63-linked chains, which enhances the intensity of the subsequent autophagy. ULK1 can be K63-linked polyubiquitinated by Tumor Necrosis Factor Receptor Associated Factor 6 (TRAF6) associated with Autophagy and Beclin1 Regulator 1 (AMBRA1), and deubiquitinated by Ubiquitin-Specific Protease 1 (USP1) (Nazio et al. 2013; Raimondi et al. 2019). The activated ULK1 complex is able to phosphorylate, thus activate Beclin1, a key subunit of the phosphatidyl-inositol-3-kinase (PtdIns3K) complex. It has been shown that Beclin1, too, can be polyubiquitinated by K63-linked chains, and this modification also affects the intensity of autophagy (Fusco et al. 2018; Shi and Kehrl 2010; Xu et al. 2016). The activated PtdIns3K produces phosphatidyl-inositol-3-phosphate that recruits Atg (autophagy-related) family proteins, which are necessary for autophagosome formation.
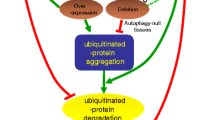
Organelles can be specifically targeted into lysosomes during selective macroautophagy. A central component of this process is the protein p62, the autophagy receptor (Lamark et al. 2017). p62 has a Ubiquitin-associated (UBA) domain that makes it able to recognise and bind to polyubiquitin, and an Microtubule-associated proteins 1A/1B light chain 3B (LC3)-Interacting Region (LIR) domain, that can bind to Atg8 (LC3 in mammals) which is localised on the phagophore membrane. A connection between the phagophore and polyubiquitinated cargo can be made through p62, the subsequent autophagosome formation and proteasome-independent degradation of the cellular component selected by ubiquitin is facilitated. This process is especially important for maintaining homeostasis in case of a dysfunctional proteasome, or for material that cannot normally be degraded by the proteasome. Most importantly, p62 is the protein that facilitates the degradation of harmful protein aggregates that are otherwise not proteasome-degradable (Lim and Lim 2011; Tan et al. 2008). The aggregated proteins get highly ubiquitinylated, with K63-linked chains, to which p62 binds and subsequently co-aggregates.
Following this, through the LIR domain and the binding of Atg8, the phagophore will be recruited, and the harmful aggregates can be sequestered from the cytoplasm. A recent finding makes this process even more intricate, as it has been shown that unanchored K48- and K63-linked polyubiquitin chains, as well a free monoubiquitin inhibits the aggregation of proteins (Zaffagnini et al. 2018).
It has been observed that during selective mitophagy, the amount of polyubiquitin is increased on the surface of the outer membrane of the damaged mitochondria, which can also be recognised by p62 (Lamark et al. 2017; Richard et al. 2020). These polyubiquitin chains are mostly assembled by the ubiquitin ligase Parkin, which is able to produce K63-linked polyubiquitin chains, too, by associating with the Ubiquitin-Conjugating Enzyme E2 13 (Ubc13) (Gao et al. 2021; Lim et al. 2013; Wang et al. 2020a). For selective mitophagy, an additional phosphorylation of ubiquitin is also essential, which is catalysed by the Phosphatase and tensin homolog INduced Kinase 1 (PINK1) ubiquitin-kinase enzyme (Lazarou et al. 2015).
During xenophagy, intracellular parasites are cleared from the cytoplasm by the lysosome, using the autophagic machinery (but like crinophagy, neither that, nor this process is considered canonical autophagy, because the degraded material does not contribute to the cell metabolism, even though it is intracellular). Multiple intracellular pathogens can be targeted for degradation by K63-linked polyubiquitination. Parasitophoric vacuoles containing type II Toxoplasma gondii are ubiquitinated by increased Interferon γ levels, mostly by K63-linked chains, which is also recognised by p62 and NDP52. In this process, the vacuoles containing pathogens are directly fusing with lysosomes, without additional autophagosome formation (Clough et al. 2016). In experiments involving Salmonella typhimurium infected cells, it has been shown that not only K63 but differently linked polyubiquitin chains are involved in the process, which can be substituted by each other (Fujita et al. 2013). The authors of this paper reached the conclusion that the ubiquitin on the Salmonella-containing vacuoles is recognised by Atg16 like 1, the ULK1 complex and Atg9 like 1.
Medical outlook
Regulation of autophagic mechanisms by K63-linked polyubiquitination has been linked to many pathological processes such as neurodegenerative and cardiovascular diseases, cancer progression, accelerated aging mechanisms and different kinds of infections (Gao et al. 2021; Lim and Lim 2011; Liu et al. 2018; Yan et al. 2018).
During the protection from neurodegenerative disorders, such as Parkinson-disease, the stabilisation of PINK1 kinase, as a positive regulator of selective macromitophagy is K63-linked polyubiquitin dependent, which is regulated by TRAF6 E3 ubiquitin ligase and its adaptor protein SARM1 (Sterile α and TIR Motif containing 1) (Murata et al. 2013). The activity of these upstream components is also essential to maintain the normal mitochondrial homeostasis and avoid neurodegenerative diseases. Moreover PKB (Protein Kinase B)/AKT, the well-known regulator of macroautophagy is also positively regulated by K63-linked polyubiquitin. The activated AKT kinase indirectly activates hypertrophic transcriptional effectors, which leads cardiac hypertrophy (Thapa et al. 2015). In evidence, autophagy has an equivalent role of the maintaining the cellular homeostatic equilibrium, in which the regulation by K63-linked polyubiquitin is essential.
Importantly, the ubiquitination code is a complex pattern, which contains several types of polyubiquitin such as the K63-K48 branched ubiquitin chains. These diverse ubiquitination pattern send different messages for the cell, so the K63-linked polyubiquitin is an important part of the branched (whole) ubiquitin pattern, which is necessary for the regulation of several types of cell biological processes (Nakasone et al. 2013).
Taken together, the role of Lys63-linked polyubiquitin is indispensable during autophagic and heterophagic processes, which are requisites of the avoiding several types of diseases.
Conclusions and future perspectives
In this review, we presented the main functions of the most important K63-linked polyubiquitin in self-digesting mechanisms. Protein ubiquitination, as a multi-functional reversible post-translational modification affects all cellular mechanisms. Moreover, autophagic processes are essential in maintaining normal cellular homeostasis via degrading and recycling unnecessary or damaged self-material from the cytoplasm. Autophagy has a colourful appearance in cells depending on the type of degraded cellular material. Interestingly, K63-linked polyubiquitin, as a frequent posttranslational signalling molecule, has a multiple role in the regulation of self-digesting processes (mostly activate or enhance autophagy). Moreover, the target organelles, protein aggregates and intracellular bacteria are designated by K63-linked ubiquitin for degradation. This phenomenon strongly reflects to the tight connection between the endo-lysosomal and ubiquitination system. Consequently, autophagic and ubiquitination processes have high medical relevance. The investigation of the connection between lysosome-dependent degradative pathways and their K63-linked polyubiquitin-mediated regulation is necessary for a better understanding of the pathological background of several self-digestion connected disorders. Therefore, it is very important to study further the role of K63-linked polyubiquitin in several types of autophagic mechanisms, which may provide the opportunity to develop new therapies and drugs for more efficient treatment of lysosome dependent self-degradative pathways related disorders.
References
Ahlberg J, Beije B, Berkenstam A, Henell F, Glaumann H (1987) Effects on in vivo and in vitro administration of vinblastine on the perfused rat liver–identification of crinosomes. Exp Mol Pathol 47:309–326
Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P (2015). Protein Function. In: Molecular Biology of the Cell. B Alberts (Eds). Garland Science. 157–161
Badarudeen B, Anand U, Mukhopadhyay S, Manna TK (2021) Ubiquitin signaling in the control of centriole duplication. FEBS J. https://doi.org/10.1111/febs.16069
Bhattacharjee A, Szabó Á, Csizmadia T, Laczkó-Dobos H, Juhász G (2019) Understanding the importance of autophagy in human diseases using Drosophila. J Genet Genomics 46:157–169
Bhattacharyya S, Yu H, Mim C, Matouschek A (2014) Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol 15:122–133
Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33:275–286
Clague MJ, Urbé S (2017) Integration of cellular ubiquitin and membrane traffic systems: focus on deubiquitinases. FEBS J 284:1753–1766
Clague M, Barsukov I, Coulson J, Liu H, Rigden D, Urbé S (2013) Deubiquitylases from genes to organism. Physiol Rev 93(3):1289–1315. https://doi.org/10.1152/physrev.00002.2013
Clague M, Heride C, Urbé S (2015) The demographics of the ubiquitin system. Trends Cell Biol 25(7):417–426. https://doi.org/10.1016/j.tcb.2015.03.002
Clough B, Wright J, Pereira P, Hirst E, Johnston A, Henriques R, Frickel E (2016) K63-linked ubiquitination targets toxoplasma gondii for endo-lysosomal destruction in IFNγ-stimulated human cells. PLOS Pathog 12(11):e1006027. https://doi.org/10.1371/journal.ppat.1006027
Coux O, Zieba BA, Meiners S (2020) The proteasome system in health and disease. Adv Exp Med Biol 1233:55–100
Csizmadia T, Juhász G (2020) Crinophagy mechanisms and its potential role in human health and disease. Prog Mol Biol Transl Sci 172:239–255
Csizmadia T, Lőw P (2020) The role of deubiquitinating enzymes in the various forms of autophagy. Int J Mol Sci 21(12):4196. https://doi.org/10.3390/ijms21124196
Csizmadia T, Lőrincz P, Hegedűs K, Széplaki S, Lőw P, Juhász G (2018) Molecular mechanisms of developmentally programmed crinophagy in Drosophila. J Cell Biol 217(1):361–374. https://doi.org/10.1083/jcb.201702145
Cuervo AM, Wong E (2014) Chaperone-mediated autophagy: roles in disease and aging. Cell Res 24:92–104
Datta A, Hura G, Wolberger C (2009) The structure and conformation of Lys63 linked tetraubiquitin. J Mol Biol 392. PMID: 19664638
Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361
Denton D, Kumar S (2019) Autophagy-dependent cell death. Cell Death Differ 26(4):605–615
Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS, Hutton M, Burrows F, Petrucelli L (2007) The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest 117:648–658
Dikic I (2017) Proteasomal and autophagic degradation systems. Annu Rev Biochem 86:193–224
Eddins M, Varadan R, Fushman D, Pickart C, Wolberger C (2007) Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol 367. PMID: 17240395
Emmerich C, Ordureau A, Strickson S, Arthur J, Pedrioli P, Komander D, Cohen P (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci 110(38):15247–15252. https://doi.org/10.1073/pnas.1314715110
Erpapazoglou Z, Walker O, Haguenauer-Tsapis R (2014) Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 3:1027–1088
Ferreira J, Soares A, Ramalho J, Pereira P, Girao H (2015) K63 linked ubiquitin chain formation is a signal for HIF1A degradation by chaperone-mediated autophagy. Sci Rep. https://doi.org/10.1038/srep10210
Fiil BK, Damgaard RB, Wagner SA, Keusekotten K, Fritsch M, Bekker-Jensen S, Mailand N, Choudhary C, Komander D, Gyrd-Hansen M (2013) OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol Cell 50:818–830
Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, Haraguchi T, Guan J, Iwai K, Tokunaga F, Saito K, Ishibashi K, Akira S, Fukuda M, Noda T, Yoshimori T (2013) Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J C Biol 203(1):115–128
Fusco C, Mandriani B, Di Rienzo M, Micale L, Malerba N, Cocciadiferro D, Sjøttem E, Augello B, Squeo G, Pellico M, Jain A, Johansen T, Fimia G, Merla G (2018) TRIM50 regulates Beclin 1 proautophagic activity. Biochim Biophys Acta Mol Cell Res 1865(6):908–919
Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jäättelä M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Münz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G (2017) Molecular definitions of autophagy and related processes. EMBO J 36:1811–1836
Gao B, Yu W, Lv P,Liang X, Sun S, Zhang Y(2021) Parkin overexpression alleviates cardiac aging through facilitating K63-polyubiquitination of TBK1 to facilitate mitophagy. Biochimica et biophysica acta. Mol Basis Dis. 1867
Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L (2015) RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep 10:226–238
Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA (1975) Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acade Sci 72(1):11–15
Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, Nakaso K, Culmsee C, Berninger B, Krappmann D, Tatzelt J, Winklhofer KF (2007) Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci 27:1868–1878
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67(1):425–479. https://doi.org/10.1146/annurev.biochem.67.1.425
Hodge CD, Leo Spyracopoulos JN, Glover M (2016) Ubc13: the Lys63 ubiquitin chain building machine. Oncotarget 7(39):64471–64504. https://doi.org/10.18632/oncotarget.10948
Hoppe T (2005) Multiubiquitylation by E4 enzymes: “one size” doesn’t fit all. Trends Biochem Sci 30:183–187
Jin J, Xie X, Xiao Y, Hu H, Zou Q, Cheng X, Sun SC (2016) Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat Immunol 17:259–268
Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635–644
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81(1):203–229. https://doi.org/10.1146/annurev-biochem-060310-170328
Komander D, Clague M, Urbé S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10(8):550–563
Komander D, Reyes‐Turcu F, Licchesi J, Odenwaelder P, Wilkinson K, Barford D (2009) Molecular discrimination of structurally equivalent Lys 63‐linked and linear polyubiquitin chains. EMBO Rep 10(6):662–662. https://doi.org/10.1038/embor.2009.106
Kong JH, Young CB, Pusapati GV, Patel CB, Ho S, Krishnan A, Lin JI, Devine W, Moreau de Bellaing A, Athni TS, Aravind L, Gunn TM, Lo CW, Rohatgi R (2020) A membrane-tethered ubiquitination pathway regulates hedgehog signaling and heart development. Dev Cell 55:432-449.e412
Kudriaeva AA, Belogurov AA (2019) Proteasome: a nanomachinery of creative destruction. Biochemistry 84:S159–S192
Kulathu Y, Komander D (2012) Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 13(8):508–523
Kwon YT, Ciechanover A (2017) The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci 42:873–886
Lamark T, Svenning S, Johansen T (2017) Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem 61(6):609–624. https://doi.org/10.1042/EBC20170035
Lazarou M, Sliter D, Kane L, Sarraf S, Wang C, Burman J, Sideris D, Fogel A, Youle R (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524(7565):309–314. https://doi.org/10.1038/nature14893
Lenoir JJ, Parisien JP, Horvath CM (2021) Immune regulator LGP2 targets Ubc13/UBE2N to mediate widespread interference with K63 polyubiquitination and NF-κB activation. Cell Rep 37:110175
Li M, Rong Y, Chuang YS, Peng D, Emr SD (2015) Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell 57:467–478
Lim K, Lim G (2011) K63-linked ubiquitination and neurodegeneration. Neurobiol Dis 43(1):9–16. https://doi.org/10.1016/j.nbd.2010.08.001
Lim G, Chew K, Ng X, Henry-Basil A, Sim R, Tan J, Chai C, Lim K (2013) Proteasome inhibition promotes Parkin-Ubc13 interaction and lysine 63-linked ubiquitination. PLoS ONE 8(9):e73235. https://doi.org/10.1371/journal.pone.0073235
Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT (2002) The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell 111:209–218
Liu P, Gan W, Siyuan S, Hauenstein AV, Tian-min F, Brasher B, Schwerdtfeger C, Liang AC, Ming X, Wei W (2018) K63-linked polyubiquitin chains bind to DNA to facilitate DNA damage repair. Sci Signal. https://doi.org/10.1126/scisignal.aar8133
Lőw P, Varga Á, Pircs K, Nagy P, Szatmári Z, Sass M, Juhász G (2013) Impaired proteasomal degradation enhances autophagy via hypoxia signaling in Drosophila. BMC Cell Biol 14:29
Marzella L, Ahlberg J, Glaumann H (1981) Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Arch B Cell Pathol Incl Mol Pathol 36:219–234
Meyer H, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157(4):910–921. https://doi.org/10.1016/j.cell.2014.03.037
Mijaljica D, Prescott M, Devenish R (2011) Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7(7):673–682. https://doi.org/10.4161/auto.7.7.14733
Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, Myung K (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci U S A 105:12411–12416
Murata H, Sakaguchi M, Kataoka K, Huh NH (2013) SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol Biol Cell 24:2772–2784
Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D (2013) Mixed-linkage ubiquitin chains send mixed messages. Structure 21:727–740
Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia G, Cecconi F (2013) mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nature Cell Biol 15(4):406–416
Nelson DL, Cox MM (2017) Peptides and Proteins. In Lehninger Principles of Biochemistry, W.H Freeman, New York
Nelson DL, Cox MM (2017b) Phosphoryl Group Transfers and ATP. Lehninger Principles of Biochemistry. W.H. Freeman, New York, pp 496–500
Nelson DL, Cox MM (2017c) Protein Metabolism. Lehninger Principles of Biochemistry. W.H. Freeman, New York, pp 1075–1077
Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, Wells JA, Gygi SP, Schulman BA, Harper JW (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell 56:360–375
Parzych K, Klionsky D (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20(3):460–473
Peng D, Zeng M, Muromoto R, Matsuda T, Shimoda K, Subramaniam M, Spelsberg T, Wei W, Venuprasad K (2011) Noncanonical K27-linked polyubiquitination of TIEG1 regulates Foxp3 expression and tumor growth. J Immunol 186(10):5638–5647
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70(1):503–533. https://doi.org/10.1146/annurev.biochem.70.1.503
Radley EH, Long J, Gough KC, Layfield R (2019) The ‘dark matter’ of ubiquitin-mediated processes: opportunities and challenges in the identification of ubiquitin-binding domains. Biochem Soc Trans 47(6):1949–1962. https://doi.org/10.1042/BST20190869
Raimondi M, Cesselli D, Di Loreto C, La Marra F, Schneider C, Demarchi F (2019) USP1 (ubiquitin specific peptidase 1) targets ULK1 and regulates its cellular compartmentalization and autophagy. Autophagy 15(4):613–630. https://doi.org/10.1080/15548627.2018.1535291
Richard T, Herzog L, Vornberger J, Rahmanto A, Sangfelt O, Salomons F, Dantuma N (2020) K63-linked ubiquitylation induces global sequestration of mitochondria. Sci Rep. https://doi.org/10.1038/s41598-020-78845-7
Rieser E, Cordier S, Walczak H (2013) Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci 38(2):94–102. https://doi.org/10.1016/j.tibs.2012.11.007
Ruocco N, Costantini S, Costantini M (2016) Blue-print autophagy: potential for cancer treatment. Mar Drugs 14(7):138. https://doi.org/10.3390/md14070138
Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455:358–362
Schulman BA, Harper JW (2009) Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol 10:319–331
Shi C, Kehrl J (2010) TRAF6 and A20 Regulate lysine 63–linked ubiquitination of beclin-1 to control TLR4-induced autophagy. Sci Signal. https://doi.org/10.1126/scisignal.2000751
Sjoerd JL, Wijk HT, Timmers M (2010) The family of ubiquitin‐conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J 24(4):981–993. https://doi.org/10.1096/fj.09-136259
Skaug B, Jiang X, Chen Z (2009) The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem 78(1):769–796. https://doi.org/10.1146/annurev.biochem.78.070907.102750
Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26:399–422
Tan J., Wong E, Kirkpatrick D, Pletnikova O, Ko H, Tay S, Ho M, Troncoso J, Gygi S, Lee M, Dawson V, Dawson T, Lim K (2008) Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Human Mol Genetics 17(3):431–439. https://doi.org/10.1093/hmg/ddm320
Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW (2009) ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol 19:1255–1263
Thapa N, Choi S, Tan X, Wise T, Anderson RA (2015) Phosphatidylinositol phosphate 5-Kinase Iγ and phosphoinositide 3-kinase/Akt signaling couple to promote oncogenic growth. J Biol Chem 290:18843–18854
Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL (2009) Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J 28:372–382
Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424:793–796
Vina-Vilaseca A, Sorkin A (2010) Lysine 63-linked polyubiquitination of the dopamine transporter requires WW3 and WW4 domains of Nedd4-2 and UBE2D ubiquitin-conjugating enzymes. J Biol Chem 285:7645–7656
Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y (2005) Chromosome alignment and segregation regulated by ubiquitination of survivin. Science 310:1499–1504
Wang X, Feng S, Wang Z, Yuan Y, Chen N, Zhang Y (2020) Parkin, an E3 ubiquitin ligase, plays an essential role in mitochondrial quality control in parkinson’s disease. Cell Mol Neurobiol 41(7):1395–1411
Weckman A, Di Ieva A, Rotondo F, Syro LV, Ortiz LD, Kovacs K, Cusimano MD (2014) Autophagy in the endocrine glands. J Mol Endocrinol 52:R151-163
Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699
Wickliffe KE, Williamson A, Meyer HJ, Kelly A, Rape M (2011) K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol 21:656–663
Xin W, Lei C, Xia T, Zhong X, Yang Q, Shu H (2019) Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat Commun. https://doi.org/10.1038/s41467-019-12145-1
Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, Li X, Liang W, Lu X, Qian L, Yuan J (2016) USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev 30(15):1718–1730
Xue B, Li H, Guo M, Wang J, Yan X, Zou X, Deng R, Li G, Zhu H (2018) TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J Virol. https://doi.org/10.1128/JVI.00321-18
Yan K, Ponnusamy M, Xin Y, Wang Q, Li P, Wang K (2018) The role of K63-linked polyubiquitination in cardiac hypertrophy. J Cell Mol Med 22:4558–4567
Yin Q, Han T, Fang B, Zhang G, Zhang C, Roberts E, Izumi V, Zheng M, Jiang S, Yin X, Kim M, Cai J, Haura E, Koomen J, Smalley K, Wan L (2019) K27-linked ubiquitination of BRAF by ITCH engages cytokine response to maintain MEK-ERK signaling. Nat Commun. https://doi.org/10.1038/s41467-019-09844-0
Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, Kuo JC, Liu CH, Lin MY, Xu M, Chen ZJ, Chen RH (2014) K33-linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell 54:586–600
Yutao W, Wen H, Jiajun F, Cai L, Li P, Zhao C, Dong Z et al (2020) Hepatocyte TNF receptor-associated factor 6 aggravates hepatic inflammation and fibrosis by promoting lysine 6–linked polyubiquitination of apoptosis signal-regulating kinase 1. Hepatology 71(1):93–11
Zaffagnini G, Savova A, Danieli A, Romanov J, Tremel S, Ebner M, Peterbauer T, Sztacho M, Trapannone R, Tarafder A, Sachse C, Martens S (2018) p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. https://doi.org/10.15252/embj.201798308
Zhang Q, Lenardo M, Baltimore D (2017) 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell 168(1–2):37–57. https://doi.org/10.1016/j.cell.2016.12.012
Zheng N, Shabek N (2017) Ubiquitin Ligases: structure, function, and regulation. Ann Rev Biochem 86:129–157
Funding
Open access funding provided by Eötvös Loránd University. This work was supported by the National Research, Development, and Innovation Office of Hungary (grant PD135447 to T. Csizmadia), the New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development, and Innovation Fund (ÚNKP-21–4-II-ELTE-60 to T. Csizmadia) and the János Bolyai Research Scholarship of the Hungarian Academy of Sciences to T. Csizmadia.
Author information
Authors and Affiliations
Contributions
AD and TC contributed equally to the planning and writing of the manuscript. AD designed the figures. All authors have read and agree to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing financial interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dósa, A., Csizmadia, T. The role of K63-linked polyubiquitin in several types of autophagy. BIOLOGIA FUTURA 73, 137–148 (2022). https://doi.org/10.1007/s42977-022-00117-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-022-00117-4