Abstract
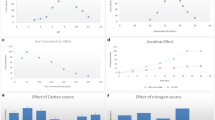
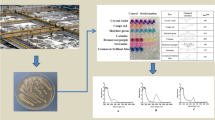
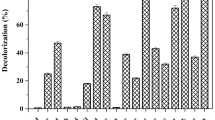
This study proposes the treatment and valorization of denim textile effluents through a fermentative hydrogen production process. Also, the study presents the decolorizing capabilities of bacterial and fungal isolates obtained from the fermented textile effluents. The maximum hydrogen production rate was 0.23 L H2/L-d, achieving at the same time color removal. A total of thirty-five bacteria and one fungal isolate were obtained from the fermented effluents and screened for their abilities to decolorize indigo dye, used as a model molecule. From them, isolates identified as Bacillus BT5, Bacillus BT9, Lactobacillus BT20, Lysinibacillus BT32, and Aspergillus H1T showed notable decolorizing capacities. Lactobacillus BT20 reached 90% of decolorization using glucose as co-substrate after 11 days of incubation producing colorless metabolites. Bacillus BT9 was able to utilize the indigo dye as the sole carbon source achieving a maximum decolorization of 60% after 9 days of incubation and producing a red-colored metabolite. In contrast, Bacillus BT5 and Lysinibacillus BT32 exhibited the lowest percentages of decolorization, barely 33% after 16 and 11 days of incubation, respectively. When Aspergillus H1T was grown in indigo dye supplemented with glucose, 96% of decolorization was reached after 2 days. This study demonstrates the valorization of denim textile effluents for the production of hydrogen via dark fermentation with concomitant color removal.




Similar content being viewed by others
References
Comisión Nacional del Agua (CONAGUA) (2011) Situación del subsector de agua potable, alcantarillado y saneamiento, Edición 2011 (situation of the subsector of drinking water, sewerage and sanitation, edition 2011), Mexico
Verma AK, Dash RR, Bhunia P (2012) A review on chemical coagulation/flocculation technologies for removal of colour from textile wastewaters. J Environ Manag 93(1):154–168
Rai HS, Bhattacharyya MS, Singh J, Bansal TK, Vats P, Banerjee UC (2005) Removal of dyes from the effluent of textile and dyestuff manufacturing industry: a review of emerging techniques with reference to biological treatment. Crit Rev Environ Sci Technol 35(3):219–238
Gomaa OM, Momtaz OA, El Kareem HA, Fathy R (2011) Isolation, identification, and biochemical characterization of a brown rot fungus capable of textile dye decolorization. World J Microbiol Biotechnol 27(7):1641–1648
Gajera HP, Bambharolia RP, Hirpara DG, Patel SV, Golakiya BA (2015) Molecular identification and characterization of novel Hypocrea koningii associated with azo dyes decolorization and biodegradation of textile dye effluents. Process Saf Environ 98:406–416
Sahasrabudhe MM, Saratale RG, Saratale GD, Pathade GR (2014) Decolorization and detoxification of sulfonated toxic diazo dye CI Direct Red 81 by Enterococcus faecalis YZ 66. J Environ Health Sci Eng 12(1):151. https://doi.org/10.1186/s40201-014-0151-1
Wang J, Lu L, Feng F (2017) Improving the indigo carmine decolorization ability of a Bacillus amyloliquefaciens laccase by site-directed mutagenesis. Cat 7(9):275
Neetha JN, Ujwal P, Sandesh K, Santhosh H, Girish K (2018) Aerobic biodegradation of acid blue-9 dye by Bacillus fermus isolated from Annona reticulata. Environ Technol Innov 11:253–261
Pensupa N, Leu SY, Hu Y, Du C, Liu H, Jing H, Wang H, Sze C, Lin K (2017) Recent trends in sustainable textile waste recycling methods: current situation and future prospects. Top Curr Chem 375(5):76
Lay CH, Kuo SY, Sen B, Chen CC, Chang JS, Lin CY (2012) Fermentative biohydrogen production from starch-containing textile wastewater. Int J Hydrogen Energ 37(2):2050–2057
Li YC, Chu CY, Wu SY, Tsai CY, Wang CC, Hung CH, Lin CY (2012) Feasible pretreatment of textile wastewater for dark fermentative hydrogen production. Int J Hydrogen Energ 37(20):15511–15517
Lin CY, Chiang CC, Nguyen TML, Lay CH (2017) Enhancement of fermentative biohydrogen production from textile desizing wastewater via coagulation-pretreatment. Int J Hydrogen Energ 42(17):12153–12158
Lin CY, Chiang CC, Nguyen TML, Lay CH (2017) Continuous biohydrogen production from coagulation-pretreated textile desizing wastewater. Int J Hydrogen Energ 42(49):29159–29165
Pérez-Rangel M, Quiroz-Figueroa FR, González-Castañeda J, Valdez-Vazquez I (2015) Microscopic analysis of wheat straw cell wall degradation by microbial consortia for hydrogen production. Int J Hydrogen Energ 40(1):151–160
Valdez-Vazquez I, Poggi-Varaldo HM (2009) Hydrogen production by fermentative consortia. Renew Sust Energ Rev 13(5):1000–1013
Valdez-Vazquez I, Sparling R, Risbey D, Rinderknecht-Seijas N, Poggi-Varaldo HM (2005) Hydrogen generation via anaerobic fermentation of paper mill wastes. Bioresour Technol 96(17):1907–1913
Lara-Vázquez A R, Sánchez A, Valdez-Vazquez I (2014) Hydration treatments increase the biodegradability of native wheat straw for hydrogen production by a microbial consortium. Int J Hydrogen Energy39(35):19899–19904
Ramya M, Anusha B, Kalavathy S (2008) Decolorization and biodegradation of indigo carmine by a textile soil isolate Paenibacillus larvae. Biodegradation 19(2):283–291
Miller GL (1959) Use of determination of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Burbano E, Sierra S, Torres K, Mercado M, Carrascal A, Poutou R (2006) Rapid DNA extraction and PCR validation for direct detection of Listeria monocytogenes in raw milk. Rev MVZ Córdova11(1):715–724
Yoon JH, Lee ST, Kim SB, Kim WY, Goodfellow M, Park YH (1997) Restriction fragment length polymorphism analysis of PCR amplified 16S ribosomal DNA for rapid identification of Saccharomonospora strains. Int J Syst Bacteriol 47(1):111–114
Luo G, Mitchell TG (2002) Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol 40(8):2860–2865
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9:678–687
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Lay CH, Sen B, Kuo SY, Chen CC, Lin CY (2014) Biohydrogen production from textile wastewater by mixed microflora in an intermittent-flow, stirred tank reactor: effect of feeding frequency. J Chin Chem Soc 61(7):791–796
Arimi MM, Knodel J, Kiprop A, Namango SS, Zhang Y, Geißen SU (2015) Strategies for improvement of biohydrogen production from organic-rich wastewater: a review. Biomass Bioenergy 75:101–118
Wong Y, Yu J (1999) Laccase-catalyzed decolorization of synthetic dyes. Water Res 33:3512–3520
Huang G, Wang W, Liu G (2015) Simultaneous chromate reduction and azo dye decolourization by Lactobacillus paracase CL1107 isolated from deep sea sediment. J Environ Manag 157:297–302
Lade H, Kadam A, Paul D, Govindwar S (2015) Biodegradation and detoxification of textile azo dyes by bacterial consortium under sequential microaerophilic/aerobic processes. EXCLI J 14:158–174
Younes SB, Sayadi S (2013) Detoxification of indigo carmine using a combined treatment via a novel trimeric thermostable laccase and microbial consortium. J Mol Catal B Enzym 87:62–68
Seesuriyachan P, Takenaka S, Kuntiya A, Klayraung S, Murakami S, Aoki K (2007) Metabolism of azo dyes by Lactobacillus casei TISTR 1500 and effects of various factors on decolorization. Water Res 41(5):985–992
Chen H, Xu H, Heinze TM, Cerniglia CE (2009) Decolorization of water and oil-soluble azo dyes by Lactobacillus acidophilus and Lactobacillus fermentum. J Ind Microbiol Biotechnol 36(12):1459–1466
Reiss R, Ihssen J, Thöny-Meyer L (2011) Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum. BMC Biotechnol 11(1):9
Lu L, Zhao M, Li GF, Li J, Wang TN, Li DB, Xu TF (2012) Decolorization of synthetic dyes by immobilized spore from Bacillus amyloliquefaciens. Catal Commun 26:58–62
Lončar N, Gligorijević N, Božić N, Vujčić Z (2014) Congo red degrading laccases from Bacillus amyloliquefaciens strains isolated from salt spring in Serbia. Int Biodeterior Biodegradation 91:18–23
Cho EA, Seo J, Lee DW, Pan G (2011) Decolorization of indigo carmine by laccase displayed on Bacillus subtilis spores. Enzym Microb Technol 49(1):100–104
Li HX, Xu B, Tang L, Zhang JH, Mao ZG (2015) Reductive decolorization of indigo carmine dye with Bacillus sp. MZS10. Int Biodeterior Biodegradation 103:30–37
Tony BD, Goyal D, Khanna S (2009) Decolorization of textile azo dyes by aerobic bacterial consortium. Int Biodeterior Biodegradation 63(4):462–469
Manu B, Chaudhari S (2003) Decolorization of indigo and azo dyes in semicontinuous reactors with long hydraulic retention time. Process Biochem 38(8):1213–1221
Balan DSL, Monteiro RTR (2001) Decolorization of textile indigo dye by ligninolytic fungi. J Biotechnol 89:141–145
Silva KML, Wanderley CRP, Marinho G, Oliveira JCD, Santos ADOD, Rodrigues K (2015) Influence of excess nitrogen in the treatment of the textile effluent in sequential batch reactors with Aspergillus niger AN 400. Eng Sanit Ambient 20(4):635–643
Khelifi E, Ayed L, Bouallagui H, Touhami Y, Hamdi M (2009) Effect of nitrogen and carbon sources on indigo and Congo red decolourization by Aspergillus alliaceus strain 121C. J Hazard Mater 163:1056–1062
Parshetti GK, Kalme SD, Gomare SS, Govindwar SP (2007) Biodegradation of reactive blue-25 by Aspergillus ochraceus NCIM-1146. Bioresour Technol 98(18):3638–3642
Jin X, Liu G, Xu Z, Wen Y (2007) Decolorization of a dye industry effluent by Aspergillus fumigatus XC6. Appl Microbiol Biotechnol 74:239–243
Kaushik P, Malik A (2011) Process optimization for efficient dye removal by Aspergillus lentulus FJ172995. J Hazard Mater 185(2–3):837–843
Abrão FO, Duarte ER, Pessoa MS, Santos VL, Freitas Júnior LF, Barros KO, Hughes AFDS, Silva TD, Rodriguez NM (2017) Notable fibrolytic enzyme production by Aspergillus spp. isolates from the gastrointestinal tract of beef cattle fed in lignified pastures. PLoS One 12(8):e0183628
Funding
This work was supported by the PROMEP/103.5/12/3680 and UGto-DAIP 000105/11. K.M. Muñoz-Páez acknowledges the support from CONACYT through the CÁTEDRAS program (Researcher ID 6407, Project 265).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Cyntia Canedo Silva
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
- Hydrogen production and decolorization of denim effluents were achieved.
- Four bacteria and one fungus were isolated from the fermented effluent.
- Lactobacillus BT20 removed indigo dye completely using glucose as co-substrate.
- Bacillus BT9 used the dye as the sole carbon source and removed 60% of color.
- Aspergillus H1T decolorized indigo dye faster than the bacteria isolated.
Rights and permissions
About this article
Cite this article
Valdez-Vazquez, I., Robledo-Rizo, J.G., Muñoz-Páez, K.M. et al. Simultaneous hydrogen production and decolorization of denim textile wastewater: kinetics of decolorizing of indigo dye by bacterial and fungal strains. Braz J Microbiol 51, 701–709 (2020). https://doi.org/10.1007/s42770-019-00157-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00157-4