Abstract
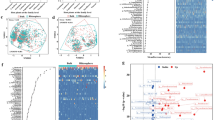
Mangroves are dynamic and unique ecosystems that provide important ecological services to coastal areas. The phylloplane is one of the greatest microbial habitats, and most of its microorganisms are uncultivated under common laboratory conditions. Bacterial community structure of Laguncularia racemosa phylloplane, a well-adapted mangrove species with salt exudation at foliar levels, was accessed through 16S rRNA amplicon sequencing. Sampling was performed in three different sites across a transect from upland to the seashore in a preserved mangrove forest located in the city of Cananéia, São Paulo State, Brazil. Higher bacterial diversity was observed in intermediary locations between the upland and the seashore, showing that significant intraspecific spatial variation in bacterial communities exists between a single host species with the selection of specific population between an environmental transect.







Similar content being viewed by others
References
Holguin G, Guzman MA, Bashan Y (1992) Two new nitrogen-fixing bacteria from the rhizosphere of mangrove trees : their isolation , identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol Ecol 101(3):207–216
Baskaran R, Mohan P, Sivakumar K, Ragavan P, Sachithanandam V (2012) Phyllosphere microbial populations of ten true mangrove species of the Andaman Island. Int J Microbiol Res [Internet] 3(26):124–127 Available from: http://idosi.org/ijmr/ijmr3(2)12/8.pdf
Holguin G, Vazquez P, Bashan Y (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fertil Soils 33(4):265–278
Wen M, Lin X, Xie M, Wang Y, Shen X, Liufu Z, Wu CI, Shi S, Tang T (2016) Small RNA transcriptomes of mangroves evolve adaptively in extreme environments. Sci Rep [Internet] 6:1–12. Available from:. https://doi.org/10.1038/srep27551
Sobrado MA (2004) Influence of external salinity on the osmolality of xylem sap , leaf tissue and leaf gland secretion of the mangrove Laguncularia racemosa ( L .). Gaertn Trees 18(4):422–427
Iermak I, Vink J, Bader AN, Wientjes E, Van Amerongen H (2016) Visualizing heterogeneity of photosynthetic properties of plant leaves with two-photon fluorescence lifetime imaging microscopy. Biochim Biophys Acta - Bioenerg 1857(9):1473–1478. Available from: https://doi.org/10.1016/j.bbabio.2016.05.005
Díaz S, Kattge J, Cornelissen JHC, Wright IJ, Lavorel S, Dray S, Reu B, Kleyer M, Wirth C, Colin Prentice I, Garnier E, Bönisch G, Westoby M, Poorter H, Reich PB, Moles AT, Dickie J, Gillison AN, Zanne AE, Chave J, Joseph Wright S, Sheremet’ev SN, Jactel H, Baraloto C, Cerabolini B, Pierce S, Shipley B, Kirkup D, Casanoves F, Joswig JS, Günther A, Falczuk V, Rüger N, Mahecha MD, Gorné LD (2015) The global spectrum of plant form and function. Nature [Internet] 529(7585):1–17. Available from:. https://doi.org/10.1038/nature16489
Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P (2017) Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol [Internet] 55(1):565–589. Available from:. https://doi.org/10.1146/annurev-phyto-080516-035623
Vorholt JA (2012) Microbial life in the phyllosphere. Nat Publ Gr [Internet] 10(12):828–840. Available from:. https://doi.org/10.1038/nrmicro2910
Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere MINIREVIEW microbiology of the phyllosphere. Appl Environ Microbiol 69(4):1875–1883
Bringel F (2015) Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front Microbiol 6(May):1–14
Ryffel F, Helfrich EJN, Kiefer P, Peyriga L, Portais JC, Piel J, Vorholt JA (2016) Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J 10(3):632–643
Karamanoli K, Menkissoglu-Spiroudi U, Bosabalidis AM, Vokou D, Constantinidou HIA (2005) Bacterial colonization of the phyllosphere of nineteen plant species and antimicrobial activity of their leaf secondary metabolites against leaf associated bacteria. Chemoecology. 15(2):59–67
Laforest-Lapointe I, Messier C, Kembel SW (2016) Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome [Internet] 4:1–10. Available from:. https://doi.org/10.1186/s40168-016-0174-1
Kembel SW, Connor TKO, Arnold HK, Hubbell SP, Wright SJ (2014) Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. PNAS. 111(38):13715–13720
Lambais MR, Crowley DE, Cury JC, Bull RC, Rodrigues RR (2006) American Association for the Advancement of Science. Science (80- ) 312(5782):18–19
Müller T, Ruppel S (2014) Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol Ecol 87(1):2–17
Peel MC, Finlayson BL, Mcmahon TA (2007) Updated world Koppen-Geiger climate classification map. Hydrol Earth Syst Sci 11:1633–1644
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”;. Proc Natl Acad Sci U S A [Internet] 2006;103(32):12115–12120. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16880384%5Cn, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1524930
Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NFY, Zhou HW (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of Illumina tags. Appl Environ Microbiol 78(23):8264–8271
Moitinho MA, Bononi L, Souza DT, Melo IS, Taketani RG (2018) Bacterial succession decreases network complexity during plant material decomposition in mangroves. Microb Ecol 76(4):954–963
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) Correspondence QIIME allows analysis of high- throughput community sequencing data intensity normalization improves color calling in SOLiD sequencing. Nat Publ Gr [Internet] 7(5):335–336. Available from:. https://doi.org/10.1038/nmeth0510-335
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing [Internet], Vienna, Austria Available from: http://www.r-project.org
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217
Love MI, Anders S, Huber W (2014) Differential analysis of count data - the DESeq2 package [Internet]. Genome Biol 15 550 p. Available from: https://doi.org/10.1101/002832%5Cnhttp://dx.doi.org/10.1186/s13059-014-0550-8
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. United Kingdom: Journal of the Royal Statiscal Society (Series B). Wiley-Blackwell; 1995
Andreote FD, Jiménez DJ, Chavez D, Dias ACF, Luvizotto DM, Dini-Andreote F et al (2012) The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One [Internet] 7(6):e38600. https://doi.org/10.1371/journal.pone.0038600 Available from: www.plosone.org
Dias AC, Franco, Taketani RG, Andreote FD, Luvizotto DM, da Silva JL et al (2012) Interspecific variation of the bacterial community structure in the phyllosphere of. Brazilian J Microbiol 43(2):653–660
Dos Santos HF, Cury JC, do Carmo FL, Dos Santos AL, Tiedje J, van Elsas JD et al (2011) Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One 6(3):1–8
Mendes LW, Tsai SM (2014) Variations of bacterial community structure and composition in mangrove sediment at different depths in Southeastern Brazil. Diversity. 6:827–843
Rigonato J, Alvarenga DO, Andreote FD, Cavalcante A, Dias F, Melo IS et al (2012) Cyanobacterial diversity in the phyllosphere of a mangrove forest. FEMS Microbiol Ecol 80:312–322
Lynch JM, Whipps JM (1990) Substrate flow in the rhizosphere. Plant Soil 129:1–10
Ruiz-Pérez CA, Restrepo S, Zambrano MM (2016) Microbial and functional diversity within the phyllosphere of Espeletia species in an Andean high-mountain ecosystem. Appl Environ Microbiol 82(6):1807–1817
Bodenhausen N, Horton MW, Bergelson J (2013) Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8(2):e56329
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B (2009) Schlapbach R. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria, PNAS
Kim M, Singh D, Lai-Hoe A, Go R, Rahim RA, Ainuddin AN et al (2012) Distinctive phyllosphere bacterial communities in tropical trees. Microb Ecol 63(3):674–681
Taketani RG, Franco NO, Rosado AS, van Elsas JD (2010) Microbial community response to a simulated hydrocarbon spill in mangrove sediments. J Microbiol [Internet] 48(1):7–15. Available from:. https://doi.org/10.1007/s12275-009-0147-1
Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N (2010) The ecology of the phyllosphere : geographic and phylogenetic variability in the distribution of bacteria. Environ Microbiol 12:2885–2893
Lovelock CE, Feller IC, McKee KL, Thompson R (2005) Variation in mangrove forest structure and sediment characteristics in Bocas del Toro. Panama Caribb J Sci 41(3):456–464
Duke NC, Ball MC, Ellison JC (1998) Factors influencing biodiversity and distributional gradients in mangroves. Glob Ecol Biogeogr Lett 7(1):27–47
Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, Loreau M, Gonzalez A (2004) The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett 7(7):601–613
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol [internet] 64:807–838 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23373698
Vokou D, Vareli K, Zarali E, Karamanoli K, Constantinidou HIA, Monokrousos N, Halley JM, Sainis I (2012) Exploring biodiversity in the bacterial community of the Mediterranean phyllosphere and its relationship with airborne bacteria. Microb Ecol 64(3):714–724
Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N (2011) Variability in the distribution of bacteria on tree leaves. Environ Microbiol 12(11):2885–2893
Gomes NCM, Borges LR, Paranhos R, Pinto FN, Leda CS (2008) Exploring the diversity of bacterial communities in sediments of urban mangrove forests. FEMS Microbiol Ecol 66(2006):96–109
Geyer W (2010) Estuarine salinity structure and circulation. In: Valle-Levinson A (ed) Contemporary Issues in Estuarine Physics (pp. 12-26). Cambridge: Cambridge University Press. https://doi.org/10.1017/CBO9780511676567.003
Cunha-Lignon M (2001) Dinâmica do manguezal no Sistema de Cananéia-Iguape, Estado de São Paulo – Brasil. Acta Bot Brasilica 15(2):56
Levin LA, Boesch DF, Covich A, Dahm C, Erséus C, Ewel KC, Kneib RT, Moldenke A, Palmer MA, Snelgrove P, Strayer D, Weslawski JM (2001) The function of marine critical transition zones and the importance of sediment biodiversity. Ecosystems. 4(5):430–451
Soares Júnior FL, Dias ACF, Fasanella CC, Taketani RG, de Sousa Lima AO, Melo IS et al (2013) Endo-and exoglucanase activities in bacteria from mangrove sediment. Brazilian J Microbiol 44(3):969–976
Bouvier TC, Del Giorgio PA (2002) Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr 47(2):453–470
Yang J, Ma L, Jiang H, Wu G, Dong H (2016) Salinity shapes microbial diversity and community structure in surface sediments of the Qinghai-Tibetan Lakes. Sci Rep [Internet] 6(1):25078 Available from: http://www.nature.com/articles/srep25078
Jiang H, Huang Q, Deng S, Dong H, Yu B (2010) Planktonic actinobacterial diversity along a salinity gradient of a river and five lakes on the Tibetan Plateau. Extremophiles. 14(4):367–376
Dillon JG, Carlin M, Gutierrez A, Nguyen V, Mclain N (2013) Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern , Baja CA Sur. Mexico Front Microbiol 4(December):1–13
Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA (2010) Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J [Internet] 4(6):719–728. Available from:. https://doi.org/10.1038/ismej.2010.9
Hunter PJ, Hand P, Pink D, Whipps JM, Bending GD (2010) Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca species) phyllosphere. Appl Environ Microbiol 76(24):8117–8125
Dini-andreote F, Stegen JC, Dirk J, Elsas V, Falcão J (2015) Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. PNAS112(11):E1326–E1332
Acknowledgments
The authors thank João Luiz da Silva from Environmental EMBRAPA for his incredible support in mangrove expeditions and samplings.
Funding
This study was financed by FAPESP’s Young Investigators grant (2013/03158-4). RGT received a Young investigator fellowship (2013/23470-2). MAM received a doctorate fellowship from CNPq. JHS received a Master’s fellowship from FAPESP (2015/23102-9).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Vania M.M. Melo.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 261 kb)
Rights and permissions
About this article
Cite this article
Moitinho, M.A., Chiaramonte, J.B., Souza, D.T. et al. Intraspecific variation on epiphytic bacterial community from Laguncularia racemosa phylloplane. Braz J Microbiol 50, 1041–1050 (2019). https://doi.org/10.1007/s42770-019-00138-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00138-7