Abstract
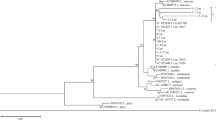
Listeria monocytogenes is one of the most important foodborne pathogens and is a causal agent of listeriosis in humans and animals. The aim of this study was to determine the prevalence, serogroups, antibiotic susceptibility, virulence factor genes, and genetic relatedness of L. monocytogenes strains isolated from 500 poultry samples in Turkey. The isolation sources of 103 L. monocytogenes strains were retail markets (n = 100) and slaughterhouses (n = 3). L. monocytogenes strains were identified as serogroups 1/2a-3a (75.7%, lineage I), 1/2c-3c (14.56%, lineage I), 1/2b-3b-7 (5.82%, lineage II), 4a-4c (2.91%, lineage III), and 4b-4d-4e (0.97%, lineage III). Most of the L. monocytogenes strains (93.2%) were susceptible to the antibiotics tested. PCR analysis indicated that the majority of the strains (95% to 100%) contained most of the virulence genes (hylA, plcA, plcB, prfA, mpl, actA, dltA, fri, flaA inlA, inlC, and inlJ). Pulsed-field gel electrophoresis (PFGE) demonstrated that there were 18 pulsotypes grouped at a similarity of > 90% among the strains. These results indicate that it is necessary to prevent the presence of L. monocytogenes in the poultry-processing environments to help prevent outbreaks of listeriosis and protect public health.




Similar content being viewed by others
References
Wehner S, Mannala GK, Qing X, Madhugiri R, Chakraborty T, Mraheil MA, Hain T, Marz M (2014) Detection of very long antisense transcripts by whole transcriptome RNA-Seq analysis of Listeria monocytogenes by semiconductor sequencing technology. PLoS One 6:e108639. https://doi.org/10.1371/journal.pone.0108639
Oxaran V, In Lee SH, Chaul LT, Corassin CH, Barancelli GV, Alves VF, de Oliveira CAF, Gram L, De Martinis ECP (2017) Listeria monocytogenes incidence changes and diversity in some Brazilian dairy industries and retail products. Food Microbiol 68:16–23. https://doi.org/10.1016/j.fm.2017.06.012
EFSA and ECDC (2018) Scientific report. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 16:5500. https://doi.org/10.2903/j.efsa.2018.5500
Doganay M (2003) Listeriosis: clinical presentation. FEMS Immunol Med Microbiol 35(3):173–175
Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV (1999) Food-related illness and death in the United States. Emerg Infect Dis 5:607–625
Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ (2014) Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. https://doi.org/10.4315/0362-028X.JFP-13-150
Todd ECD, Notermans S (2011) Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Control 22:1484–1490. https://doi.org/10.1016/j.foodcont.2010.07.021
Garner D, Kathariou S (2016) Fresh produce-associated listeriosis outbreaks, sources of concern, teachable moments, and insights. J Food Prot 79:337–344. https://doi.org/10.4315/0362-028X.JFP-15-387
Du X-J, Zhang X, Wang X-Y, Su Y-I, Li P, Wang S (2017) Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Control 74:9–16. https://doi.org/10.1016/j.foodcont.2016.11.024
Soni DK, Singh M, Singh DV, Dubey SK (2014) Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol 14:241. https://doi.org/10.1186/s12866-014-0241-3
Allen KJ, Walecka-Zacharska E, Chen JC, Katarzyna KP, Devlieghere F, Van Meervenne E, OSek J, Wieczorek K, Bania J (2016) Listeria monocytogenes: an examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol 54:178–189. https://doi.org/10.1016/j.fm.2014.08.006
Sakaridis I, Soultos N, Iossifidou E, Papa A, Ambrosiadis I, Koidis P (2011) Prevalence and antimicrobial resistance of Listeria monocytogenes isolated in chicken slaughterhouses in northern Greece. J Food Prot 74:1017–1021. https://doi.org/10.4315/0362-028X.JFP-10-545
EFSA and ECDC (2014) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J 12:3547. https://doi.org/10.2903/j.efsa.2014.3547
Burall LS, Simpson AC, Datta AR (2011) Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multiplex PCR assay for identifying the major serotypes of Listeria monocytogenes. J Food Prot 74:403–409. https://doi.org/10.4315/0362-028X
Chen JQ, Regan P, Laksanalamai P, Healey S, Hu Z (2017) Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci Human Wellness 6:97–120. https://doi.org/10.1016/j.fshw.2017.06.002
Huang Y, Morvay AA, Shi X, Suo Y, Shi C, Knøchel S (2018) Comparison of oxidative stress response and biofilm formation of Listeria monocytogenes serotypes 4b and 1/2a. Food Control 85:416–422. https://doi.org/10.1016/j.foodcont.2017.10.007
Orsi RH, den Bakker HC, Wiedmann M (2011) Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. https://doi.org/10.1016/j.ijmm.2010.05.002
Cocolin L, Stella S, Nappi R, Bozzetta E, Cantoni C, Comi G (2005) Analysis of PCR-based methods for characterization of Listeria monocytogenes strains isolated from different sources. Int J Food Microbiol 103:167–178. https://doi.org/10.1016/j.ijfoodmicro.2004.12.027
Graves LM, Swaminathan B (2001) PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol 65:55–62
Parisi A, Latorre L, Normanno G, Miccolupo A, Fraccalvieri R, Lorusso V, Santagada G (2010) Amplified fragment length polymorphism and multi-locus sequence typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food Microbiol 27:101–108. https://doi.org/10.1016/j.fm.2009.09.001
Cao X, Wang Y, Wang Y, Ye C (2018) Isolation and characterization of Listeria monocytogenes from the black-headed gull feces in Kunming, China. J Infect Public Health 11:59–63. https://doi.org/10.1016/j.jiph.2017.03.003
Dussurget O, Pizzaro-Cerda J, Cossart P (2004) Molecular determinants of Listeria monocytogenes virulence. Annu Rev Microbiol 58:587–610. https://doi.org/10.1146/annurev.micro.57.030502.090934
Ye K, Zhang X, Huang Y, Liu J, Liu M, Zhou G (2018) Bacteriocinogenic Enterococcus faecium inhibits the virulence property of Listeria monocytogenes. LWT Food Sci Technol 89:87–92. https://doi.org/10.1016/j.lwt.2017.10.028
Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, Cossart P, European Listeria Genome Consortium (2002) Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol 45:1095–1106
Bonazzi M, Lecuit M, Cossart P (2009) Listeria monocytogenes internalin and E-cadherin: from bench to bedside. Cold Spring Harb Perspect Biol 1:a003087. https://doi.org/10.1101/cshperspect.a003087
Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Vázquez-Boland JA, Kunst F, Cossart P, Buchrieser C (2003) Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47:1613–1625
Soni DK, Ghosh A, Chikara SK, Singh KM, Joshic C, Dubey SK (2017) Comparative whole genome analysis of Listeria monocytogenes 4b strains reveals least genome diversification irrespective of their niche specificity. Gene Reports 8:61–68. https://doi.org/10.1016/j.genrep.2017.05.007
ISO (2004) Microbiology of food animal feeding stuffs: Horizontal method for the detection and enumeration of Listeria monocytogenes. Part 1: detection method Amendment 1: modification of isolation media and haemolysis test, and inclusion of precision data. ISO 11290-1/A1, Geneva, Switzerland
Liu D, Ainsworth AJ, Austin FW, Lawrence ML (2004) Use of PCR primers derived from a putative transcriptional regulator gene for species-specific determination of Listeria monocytogenes. Int J Food Microbiol 91:297–304. https://doi.org/10.1016/j.ijfoodmicro.2003.07.004
Bubert A, Köhler S, Goebel W (1992) The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl Environ Microbiol 58:2625–2632
Bubert A, Sokolovic Z, Chun SK, Papatheodorou L, Simm A, Goebel W (1999) Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol Gen Genet 261:323–336
Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P (2004) Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. https://doi.org/10.1128/JCM.42.8.3819-3822.2004
EUCAST (2016) European committe on antimicrobial susceptibility testing. Breakpoints tables for interpretation of MICs and zone diameters, Version 6.0. Retrieved from http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf. Accessed 26 January 2019
Furrer B, Candrian U, Hoefelein C, Luethy J (1991) Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Microbiol 70:372–379. https://doi.org/10.1111/j.1365-2672.1991.tb02951.x
Mengaud J, Vicente MF, Cossart P (1989) Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect Immun 57:3695–3701
Nishibori T, Cooray K, Xiong H, Kawamuro I, Fujita M, Mitsuyama M (1995) Correlation between the presence of virulence-associated genes as determined by PCR and actual virulence to mice in various strains of Listeria spp. Microbiol Immunol 39:343–349
Liu D, Lawrence ML, Austin FW, Ainsworth AJ (2007) A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J Microbiol Methods 71:133–140. https://doi.org/10.1016/j.mimet.2007.08.007
Slama RB, Miladi H, Chaieb K, Bakhrouf A (2013) Survival of Listeria monocytogenes cells and the effect of extended frozen storage (−20 °C) on the expression of its virulence gene. Appl Biochem Biotechnol 170:1174–1183. https://doi.org/10.1007/s12010-013-0253-8
Jaradat ZW, Schutze GE, Bhunia AK (2002) Genetic homogeneity among Listeria monocytogenes strains from infected patients and meat products from two geographic locations determined by phenotyping, ribotyping and PCR analysis of virulence genes. Int J Food Microbiol 76:1–10
Leimeister-Wächter M, Domann E, Chakraborty T (1991) Detection of a gene encoding a phosphatidylinositol-specific phospholipase C that is coordinately expressed with listeriolysin in Listeria monocytogenes. Mol Microbiol 5:361–366
Kyoui D, Takahashi H, Miya S, Kuda T, Kimura B (2014) Comparison of the major virulence-related genes of Listeria monocytogenes in Internalin A truncated strain 36-25-1 and a clinical wild-type strain. BMC Microbiol 14:15. https://doi.org/10.1186/1471-2180-14-15
Promadej N, Fiedler F, Cossart P, Dramsi S, Kathariou S (1999) Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J Bacteriol 181:418–425
Vazquez-Boland JA, Kochs C, Dramsi S, Ohayaon H, Geoffroy C, Mengaud J, Cossart P (1992) Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun 60:219–230
Oliveira TS, Varjão LM, Silva LNN, Pereira RCL, Hofer E, Vallim DC, Almeida RCC (2018) Listeria monocytogenes at chicken slaughterhouse: occurrence, genetic relationship among isolates and evaluation of antimicrobial susceptibility. Food Control 88:131–138. https://doi.org/10.1016/j.foodcont.2018.01.015
Khallaf M, Ameur N, Boraam F, Senouci S, Ennaji MM (2016) Prevalence of Listeria monocytogenes isolated from chicken meat marketed in Rabat, Morocco. Int J Innov Sci Res 22:8–13
Bouayad L, Hamdi TM, Naim M, Leclercq A, Lecuit M (2015) Prevalence of Listeria spp. and molecular characterization of Listeria monocytogenes isolates from broilers at the abattoir. Foodborne Pathog Dis 12:606–611. https://doi.org/10.1089/fpd.2014.1904
Dan SD, Tabaran A, Mihaiu L, Mihaiu M (2015) Antibiotic susceptibility and prevalence of foodborne pathogens in poultry meat in Romania. J Infect Dev Ctries 9:35–41. https://doi.org/10.3855/jidc.4958
Kahraman T, Aydin A (2009) Prevalence of Salmonella spp., Escherichia coli O157:H7 and Listeria monocytogenes in meat and meat products in Turkey. Arch Leb 60:6–11. https://doi.org/10.2376/0003-925X-60-6
Elmali M, Can HY, Yaman H (2015) Prevalence of Listeria monocytogenes in poultry meat. Food Sci Technol 35:672–675. https://doi.org/10.1590/1678-457X.6808
Ristori CA, Rowlands RE, Martins CG, Barbosa ML, Yoshida JT, Franco BD (2014) Prevalence and populations of Listeria monocytogenes in meat products retailed in Sao Paulo, Brazil. Foodborne Pathog Dis 11:969–973. https://doi.org/10.1089/fpd.2014.1809
Amajoud N, Lecrerq A, Soriano JM, Bracq-Dieve H, El Maadoudi M, Senhaji NS, Kounnoun A, Moura A, Lecuit M, Abrini J (2018) Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Control 84:436–441. https://doi.org/10.1016/j.foodcont.2017.08.023
Vongkamjan K, Benjakul S, Vu HTK, Vuddhakul V (2017) Longitudinal monitoring of Listeria monocytogenes and Listeria phages in seafood processing environments in Thailand. Food Microbiol 66:11–19. https://doi.org/10.1016/j.fm.2017.03.014
Leong D, Alvarez-Ordóňez A, Jordon K (2014) Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front Microbiol 5:436. https://doi.org/10.3389/fmicb.2014.00436
Murugesan L, Kucerova Z, Knabel SJ, LaBorde LF (2015) Predominance and distribution of a persistent Listeria monocytogenes clone in a commercial fresh mushroom processing environment. J Food Prot 78:1988–1998. https://doi.org/10.4315/0362-028X
Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, Criscuolo A, Gaultier C, Roussel S, Brisabois A, Disson O, Rocha EPC, Brisse S, Lecuit M (2016) Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 48:308–313. https://doi.org/10.1038/ng.3501
Li R, Du W, Yang J, Liu Z, Yousef AE (2018) Control of Listeria monocytogenes biofilm by paenibacterin, a natural antimicrobial lipopeptide. Food Control 84:529–535. https://doi.org/10.1016/j.foodcont.2017.08.031
Hilliard A, Leong D, O’Callaghan A, Culligan EP, Morgan CA, De Lappe N, Hill C, Jordan K, Cormican M, Gahan CGM (2018) Genomic characterization of Listeria monocytogenes isolates associated with clinical listeriosis and the food production environment in Ireland. Genes 9:171. https://doi.org/10.3390/genes9030171
Iglesias MA, Kroning IS, Decol LT, de Melo Franco BDG, Silva WP (2017) Occurrence and phenotypic and molecular characterization of Listeria monocytogenes and Salmonella spp. in slaughterhouses in southern Brazil. Food Res Int 100:96–101. https://doi.org/10.1016/j.foodres.2017.06.023
Doménech E, Jimenez-Belenguer A, Amoros JA, Ferrus MA, Escriche I (2015) Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in eastern Spain. Food Control 47:120–125. https://doi.org/10.1016/j.foodcont.2014.06.043
Hof H (2004) An update on the medical management of listeriosis. Expert Opin Pharmacother 5:1727–1735. https://doi.org/10.1517/14656566.5.8.1727
Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, Dao TT, Courvalin P, Brisabois A (2011) Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl Environ Microbiol 77:2788–2790. https://doi.org/10.1128/AEM.01381-10
Sugiri YD, Gölz G, Meeyam T, Baumann MP, Kleer J, Chaisowwong W, Alter T (2014) Prevalence and antimicrobial susceptibility of Listeria monocytogenes on chicken carcasses in Bandung, Indonesia. J Food Prot 77:1407–1410. https://doi.org/10.4315/0362-028X.JFP-13-453
European Commission (2011) Commission recommendation on the research joint programming initiative ‘The microbial challenge-an emerging threat to human health.’ Brussels, Retrieved from https://era.gv.at/object/document/677/attach/CommRec_DOCUMENTDETRAVAIL_f.pdf. Accessed 27 January 2019
Akrami-Mohajeri F, Derakhshan Z, Ferrante M, Hamidiyan N, Soleymani M, Conti GO, Tafti RD (2018) The prevalence and antimicrobial resistance of Listeria spp. in raw milk and traditional dairy products delivered in Yazd, Central Iran (2016). Food Chem Toxicol 114:141–144. https://doi.org/10.1016/j.fct.2018.02.006
Rezai R, Ahmadi E, Salimi B (2018) Prevalence and antimicrobial resistance profile of Listeria species isolated from farmed and on-sale rainbow trout (Oncorhynchus mykiss) in Western Iran. J Food Prot 81:886–891. https://doi.org/10.4315/0362-028X.JFP-17-428
Sharma S, Sharma V, Dahiya DK, Khan A, Mathur M, Sharma A (2017) Prevalence, virulence potential, and antibiotic susceptibility profile of Listeria monocytogenes isolated from bovine raw milk samples obtained from Rajasthan, India. Foodborne Pathog Dis 14:132–140. https://doi.org/10.1089/fpd.2016.2118
OECD (2018) Stopping antimicrobial resistance would cost just USD 2 per person a year. https://www.oecd.org/newsroom/stopping-antimicrobial-resistance-would-cost-just-usd-2-per-person-a-year.htm. Accessed 13 February 2019
Walsh D, Duffy G, Sheridan JJ, Blair IS, McDowell DA (2001) Antibiotic resistance among Listeria, including Listeria monocytogenes, in retail foods. J Appl Microbiol 90:517–522
Threlfall EJ, Skinner JA, McLauchlin J (1998) Antimicrobial resistance in Listeria monocytogenes from humans and food in the UK, 1967-96. Clin Microbiol Infect 4:410–412. https://doi.org/10.1111/j.1469-0691.1998.tb00087.x
Gómez D, Azon E, Marco N, Carraminana JJ, Rota C, Ariňo A, Yangüela J (2014) Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol 42:61–65. https://doi.org/10.1016/j.fm.2014.02.017
Ayaz ND, Erol I (2010) Relation between serotype distribution and antibiotic resistance profiles of Listeria monocytogenes isolated from ground Turkey. J Food Prot 73:967–972. https://doi.org/10.4315/0362-028X-73.5.967
Byrne VV, Hofer E, Vallim DC, de Castro Almeida RC (2016) Occurrence and antimicrobial resistance patterns of Listeria monocytogenes isolated from vegetables. Braz J Microbiol 47:438–443. https://doi.org/10.1016/j.bjm.2015.11.033
Popovich KJ, Snitkin ES (2017) Whole genome sequencing-implications for infection prevention and outbreak investigations. Curr Infect Dis Rep 19:15. https://doi.org/10.1007/s11908-017-0570-0
Mendonça KS, Michael GB, von Laer AE, Menezes DB, Cardoso MRI, Silva WP (2012) Genetic relatedness among Listeria monocytogenes isolated in foods and food production chain in southern Rio Grande do Sul, Brazil. Food Control 28:171–177. https://doi.org/10.1016/j.foodcont.2012.04.014
Madden RH, Hutchison M, Jordan K, Pennone V, Gundogdu O, Corcionivoschi N (2018) Prevalence and persistence of Listeria monocytogenes in premises and products of small food business operators in Northern Ireland. Food Control 87:70–78. https://doi.org/10.1016/j.foodcont.2017.12.020
Funding sources
This study was funded by Istanbul University with the project number 56821. Additional fund was obtained from the Scientific and Technological Research Council of Turkey (TUBITAK 2214/A) to support Aysen COBAN on an International Doctoral Research Scholarship Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Luis Augusto Nero.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Coban, A., Pennone, V., Sudagidan, M. et al. Prevalence, virulence characterization, and genetic relatedness of Listeria monocytogenes isolated from chicken retail points and poultry slaughterhouses in Turkey. Braz J Microbiol 50, 1063–1073 (2019). https://doi.org/10.1007/s42770-019-00133-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-019-00133-y