Abstract
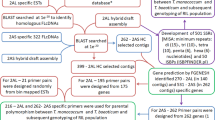
Karnal bunt of wheat is an internationally quarantined disease caused by Tilletia indica. The disease is important in North-western plain zone of India. Till date no reference or complete genome of Tilletia indica is available and reported genomes are variable in size ranging from 26.7 to 43.7 Mb. Recently whole genome of Tilletia indica RAKB_UP_1 (GCA_002220835.1, IARI, New Delhi) has been sequenced. In present study, the genomes of seven isolates of T. indica viz.. DAOM_236416 (GCA_001645015.1), PSWKBGD_1_3 (GCA_001689965.1), PSWKBGH_1 (GCA_001689995.1), PSWKBGH_2 (GCA_001689945.1), RAKB_UP_1 (GCA_002220835.1), TiK_1 (GCA_002997305.1) and Tik (GCA_003054935.1) were compared. For genome mapping, all the genome assemblies were aligned against RAKB_UP_1 genome using minimap2 wherein 86–100% similarity was observed with the reference genome. Tik_1 mapped 100%. Further, comparative genome analysis was done by identification of SNPs, In-Dels, segmental duplicates, LTR and CNV. It revealed that variable number of SNPs were ranging from 1380 (Tik_1) to 160966 (Tik) and 82 (PSWKBGH_1) to 3309 (PSWKBGD_1_3) In-Dels. The maximum number of segmental duplications (SDs) were identified in TiK_1 isolate (60632 regions) while least in DAOM_236416 isolate (2082 regions). LTR retrotransposon regions were found as 307 (DAOM_236416)–833 (PSWKBGH_2). The copy number variations including duplications and deletions have also been identified and Tik had shown highest number of duplications count (100) while no duplication was found in Tik_1 which was expected also because of 100% mapping with the reference genome. In case of deletion counts, highest (37) deletion were detected in DAOM_236416. This first study revealed that T. indica genome showed high genomic variation and can be used to devise the novel management strategies against Karnal bunt of wheat.


Similar content being viewed by others
References
Abyzov A, Urban AE, Snyder M, Gerstein M (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21(6):974–984. https://doi.org/10.1101/gr.114876.110
Agarwal VK, Verma HS (1983) A simple technique for the detection of karnal bunt infection in wheat seed samples. Seed Res 11(1):100–102
Aggarwal R, Tripathi A, Yadav A (2010) Pathogenic and genetic variability in Tilletia indica monosporidial culture lines using universal rice primer-PCR. Eur J Plant Pathol 128:333–342. https://doi.org/10.1007/s10658-010-9655-4
Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11(6):1005–1017. https://doi.org/10.1101/gr.gr-1871r
Browning SR, Browning BL (2016) Genotype imputation with millions of reference samples. Am J Hum Genet 98:116–126. https://doi.org/10.1086/j.ajhg.2015.11.020
Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6(2):80–92. https://doi.org/10.4161/fly.19695
Dhaliwal HS, Singh DV (1989) Production and inter-relationship of two types of secondary sporidia of Neovossia indica. Curr Sci 58:614–618
Duran R, Cromarty R (1977) Tilletia indica: a heterothallic wheat bunt fungus with multiple alleles controlling incompatibility. Phytopathology 67:812–815
Ellinghaus D, Kurtz S, Willhoeft U (2008) LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics14;9:18. https://doi.org/10.1186/1471-2105-9-18
Gurjar MS, Jogawat A, Kulshreshtha D, Sharma S, Gogoi R, Aggarwal R (2016) Intraspecific variation of Tilletia indica isolates causing Karnal bunt of wheat in India. Indian Phytopath 69:352–356
Gurjar MS, Aggarwal R, Sharma S, Kulshreshtha D, Gupta A, Gogoi R, Thirumalaisamy PP, Saini A (2017) Development of real time PCR assay for the detection and quantification of teliospores of Tilletia indica causing wheat Karnal bunt in soil. Indian J Exp Biol 55:549–554
Gurjar MS, Jogawat A, Saharan MS, Aggarwal R (2018) Response of putative pathogenicity-related genes in Tilletia indica inciting Karnal bunt of wheat. Cereal Res Commun 46(1):89–103. https://doi.org/10.1556/0806.45.2017.06
Gurjar MS, Aggarwal R, Jogawat A, Kulshreshtha D, Sharma S, Solanke AU, Dubey H, Jain RK (2019) Genome sequencing and secretome analysis of Tilletia indica inciting Karnal bunt of wheat provides pathogenesis-related genes. Biotech 3 9(6):219. https://doi.org/10.1007/s13205-019-1743-3
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 15(18):34. 3094–3100. https://doi.org/10.1093/bioinformatics/bty191
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) 1000 Genome project data processing subgroup. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Mishra P, Maurya R, Gupta VK, Ramteke PW, Marla SS, Kumar A (2019) Comparative genomic analysis of monosporidial and monoteliosporic cultures for unraveling the complexity of molecular pathogenesis of Tilletia indica pathogen of wheat. Sci Rep 9:8185. https://doi.org/10.1038/s41598-019-44464-0
Mitra M (1931) A new bunt on wheat in India. Ann App Biol 18(2):178–179. https://doi.org/10.1111/j.1744-7348.1931.tb02294.x
Numanagic I, Gokkaya AS, Zhang L, Berger B, Alkan C, Hach F (2018) Fast characterization of segmental duplications in genome assemblies. Bioinformatics 34(17):i706–i714. https://doi.org/10.1093/bioinformatics/bty586
Sharma P, Tiwari R, Saharan MS, Sharma I et al (2016) Draft genome sequence of two monosporidial lines of the Karnal bunt fungus Tilletia indica Mitra (PSWKBGH-1 and PSWKBGH-2). Genome Announc 4(5):e00928–e00916
Tan MK, Brennan JP, Wright D, Murray GM (2013) A review of the methodology to detect and identify Karnal bunt-a serious biosecurity threat. Australas Plant Pathol 42:95–102. https://doi.org/10.1007/s13313-012-0176-9
Tan MK, Raman H, Chambers G, Sharma I, Chen Z et al (2016) Characterization of SNP and structural variations in the mitochondrial genomes of Tilletia indica and its closely related species formed basis for a simple diagnostic assay. Plos One 11(11):e0166086
Zhao S, Gibbson JG (2018) A population genomic characterization of copy number variation in the opportunistic fungal pathogen Aspergillus fumigates. PLos One 13(8):e0201611
Zheng W, Huang L, Huang J et al (2013) High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat Commun 4:2673
Acknowledgements
This research work is financially supported by the ICAR-Consortium Research Platform (CRP) on Genomics (ICAR-G/CRP-Genomics/2015–2720). We are highly thankful to the Director, Joint Director (Research), ICAR-IARI, New Delhi for providing guidance and facilities for investigations.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gurjar, M.S., Aggarwal, R., Jain, P. et al. Comparative genome analysis of Tilletia indica inciting Karnal bunt of wheat reveals high genomic variation. Indian Phytopathology 73, 707–712 (2020). https://doi.org/10.1007/s42360-020-00260-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-020-00260-9