Abstract
Safflower wilt, incited by Fusarium oxysporum f. sp. carthami (Foc) is one of the most devastating diseases of safflower. Virulence and genetic diversity of 90 Foc isolates representing four major safflower growing states (Maharashtra, Karnataka, Telangana, and Madhya Pradesh) in India were analyzed. Out of 45 RAPD and 45 ISSR primers screened, 17 of each RAPD and ISSR markers revealed a comprehensive picture of genetic diversity. Primers depicting maximum heterozygosity, effective number of alleles, and Shannon information index were identified. Cluster and principal coordinate analyses distributed isolates from a given state among different clusters and quadrants, respectively, indicating high genetic diversity. However, certain region-specific groupings within clusters were found. The population structure comprised two sub-populations with admixture of alleles. AMOVA indicated maximum molecular variation within states (95%), and within races (79%). Among the six races identified on the basis of disease reactions on differential cultivars, Race 6 was the most virulent and Race 2 as the most prevalent. Groupings on the basis of molecular markers and geographical origin showed some degree of concurrence with the virulence of isolates. The findings are useful in screening of newly developed pathogen resistant varieties for the deployment of region-specific resistant cultivars of safflower.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215:403–410
Anand G, Kapoor R (2018) Population structure and virulence analysis of Alternaria carthami isolates of India using ISSR and SSR markers. World Journal of Microbiology and Biotechnology 34:140
Arif M, Zaidi NW, Haq QM, Singh YP, Taj G, Kar CS, Singh US (2015) Morphological and comparative genomic analyses of pathogenic and non-pathogenic Fusarium solani isolated from Dalbergia sissoo. Molecular Biology Reports 42:1107–1122
Baayen RP, O'Donnell K, Bonants PJ, Cigelnik E, Kroon LP, Roebroeck EJ, Wallwijk C (2000) Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and non-monophyletic formae speciales causing wilt and rot disease. Phytopathology 90:891–900
Bayraktar H, Dolar FS, Maden S (2008) Use of RAPD and ISSR markers in detection of genetic variation and population structure among Fusarium oxysporum f. sp. ciceris isolates on chickpea in Turkey. Journal of Phytopathology 156:146–154
Baysal Ö, Siragusa M, Gümrükcü E, Zengin S, Carimi F, Sajeva M, Da Silva JA (2010) Molecular characterization of Fusarium oxysporum f. sp. melongenae by ISSR and RAPD markers on eggplant. Biochemical Genetics 48:524–537
Bogale M, Wingfield BD, Wingfield MJ, Steenkamp ET (2005) Simple sequences repeat markers for species in the Fusarium oxysporum complex. Molecular Ecology Notes 5:622–624
Booth C (1971) The Genus Fusarium. Commonwealth Mycological Institute, Kew Surrey
Cabanás CG, Valverde-Corredor A, Pérez-Artés E (2012) Molecular analysis of Spanish populations of Fusarium oxysporum f. sp. dianthi demonstrates a high genetic diversity and identifies virulence groups in races 1 and 2 of the pathogen. European Journal of Plant Pathology 132:561–576
Costa SN, Bragança CA, Ribeiro LR, Amorim EP, Oliveira SA, Dita MA, Laranjeira FF, Haddad F (2014) Genetic structure of Fusarium oxysporum f. sp. cubense in different regions from Brazil. Plant Pathology 64:137–146
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dubey SC, Singh SR (2008) Virulence analysis and oligonucleotide fingerprinting to detect diversity among Indian isolates of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Mycopathologia 165:389–406
Dubey SC, Tripathi A, Singh SR (2010) ITS-RFLP fingerprinting and molecular marker for detection of Fusarium oxysporum f. sp. ciceris. Folia Microbiologica 55:629–634
Earl DA, VonHoldt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
FAOSTAT (2015) Food and Agriculture Organization of the United Nations, FAO Statistical Databases Agricultural production data; Available: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor. Accessed 3 Jan 2017.
Gale LR, Katan T, Kistler HC (2003) The probable center of origin of Fusarium oxysporum f. sp. lycopersici VCG 0033. Plant Disease 87:1433–1438
Gilbert J (2008) International safflower production - an overview. In: Knights SE, Potter TD (eds) Safflower: unexploited potential and world adaptability, proceedings of the 7th international safflower conference. Australian Oilseeds Federation. Wagga Wagga, Australia
Gordon TR, Martyn RD (1997) The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology 35:111–128
Groenewald S, Van Den Berg N, Marasas WF, Viljoen A (2006) The application of high-throughput AFLP's in assessing genetic diversity in Fusarium oxysporum f. sp. cubense. Fungal Biology 110:297–305
Gupta SK (2015) Breeding oilseed crops for sustainable production: opportunities and constraints. Academic Press, Cambridge
Haware MP, Nene YL (1982) Races of Fusarium oxysporum f. sp. ciceri. Plant Disease 66:809–810
Haware MP, Nene YL, Natarajan M (1996) The survival of Fusarium oxysporum f. sp. ciceri in the soil in the absence of chickpea. Phytopathologia Mediterranea 35:9–12
Henrique FH, Carbonell SA, Ito MF, Gonçalves JG, Sasseron GR, Chiorato AF (2015) Classification of physiological races of Fusarium oxysporum f. sp. phaseoli in common bean. Bragantia 74:84–92
Indiastat (2015) https://www.indiastat.com/agriculture/2/oilseeds/17204/safflower/19580/stats.aspx. Accessed 3 Jan 2017.
Kalpana Sastry R (1996) Symptoms of wilt disease clues for use in resistance breeding. In: Hegde DM, Raghavaiah CV, Pati D (eds) Proceedings of training program on breeding approaches for improving productivity of safflower and group meeting on heterosis breeding in safflower. Directorate of Oilseed Research, Rajendranagar, Hyderabad, pp 25–32
Kalpana Sastry R, Chattopadhyay C (2003) Development of Fusarium wilt resistant genotypes in safflower (Carthamus tinctorius). European Journal of Plant Pathology 109:147–151
Kandan A, Akhtar J, Singh B, Dev U, Goley R, Chand D, Roy A, Rajkumar S, Agarwal PC (2014) Genetic diversity analysis of Alternaria alternata isolates infecting different crops using URP and ISSR markers. Indian Journal of Plant Protection 42:229–236
Kandan A, Akhtar J, Singh B, Pal D, Chand D, Rajkumar S, Agarwal P (2016) Genetic diversity analysis of fungal pathogen Bipolaris sorghicola infecting Sorghum bicolor in India. Journal of Environmental Biology 37:1323–1330
Klisiewicz JM (1975) Race 4 of Fusarium oxysporum f. sp. carthami. Plant Disease Reporter 59:712–714
Klisiewicz JM, Houston BR (1962) Fusarium wilt of safflower. Plant Disease Reporter 46:748–749
Klisiewicz JM, Thomas CA (1970) Pathogenic races of Fusarium oxysporum f. sp. carthami. Phytopathology 60:83–84
Liu N, Liu ZL, Gong G, Zhang M, Wang X, Zhou Y, Qi X, Chen H, Yang J, Luo P, Yang C (2015) Virulence structure of Blumeria graminis f. sp. tritici and its genetic diversity by ISSR and SRAP profiling analyses. PLoS One 10:e0130881
Mahfooz S, Maurya DK, Srivastava AK, Kumar S, Arora DK (2012) A comparative in silico analysis on frequency and distribution of microsatellites in coding regions of three formae speciales of Fusarium oxysporum and development of EST–SSR markers for polymorphism studies. FEMS Microbiology Letters 328:54–60
Majidi MM, Tavakoli V, Mirlohi A, Sabzalian MR (2011) Wild safflower species (Carthamus oxyacanthus Bieb.): a possible source of drought tolerance for arid environments. Australasian Journal of Crop Science 5:1055–1063
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Research 27:209–220
McDonald BA (1997) The population genetics of fungi: tools and techniques. Phytopathology 87:448–453
McDonald BA, Linde C (2002) The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124:163–180
Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, Powell W, Waugh R (1997) Comparison of PCR based marker systems for the analysis of genetic relationships in cultivated potato. Molecular Breeding 3:127–136
Murumkar DR (2010) Variability among isolates of Fusarium oxysporum f. sp. carthami from Madhya Pradesh. Indian Phytopathology 63:446–448
Murumkar DR, Indi DV, Gud MA, Deshpande AN (2008) Variability among isolates of Fusarium oxysporum f. sp. carthami from Maharashtra state. Journal of Plant Disease Sciences 3:24–28
Nelson PE, Toussoun TA, Marasas WFO (1983) Fusarium species. An Illustrated Manual for Identification. Pennsylvania State University Press, University Park, p 193
Nene YL, Haware MP, Reddy MV (1981) Chickpea diseases: resistance - screening techniques. International crop research Institute for the Semi-Arid Tropics, Patancheru, A. P, India
Nirmaladevi D, Venkataramana M, Srivastava RK, Uppalapati SR, Gupta VK, Yli-Mattila T, Tsui KC, Srinivas C, Niranjana SR, Chandra NS (2016) Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Scientific Reports 6:21367–21381
Öztürk E, Özer H, Polat T (2008) Growth and yield of safflower genotypes grown under irrigated and non-irrigated conditions in a highland environment. Plant Soil and Environment 54:453–460
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research - an update. Bioinformatics 28:2537–2539
Perrier X, Jacquemoud-Collet JP (2006) DARwin software: dissimilarity analysis and representation for windows. http://darwin.Cirad.Fr/Darwin. Accessed 11 Mar 2017.
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical and Applied Genetics 98:107–112
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raghuwanshi KS, Dake GN, Mate SN, Naik RM (2008) Pathogenic variations of Fusarium oxysporum f. sp. carthami. Journal of Plant Disease Sciences 3:241–242
Rao VN, Sastry RK, Craufurd P, Meinke H, Parsons D, Rego TJ, Rathore A (2014) Cropping systems strategy for effective management of Fusarium wilt in safflower. Field Crops Research 156:191–198
Sato T, Sugawara K, Kudo N (2008) Occurrence of Fusarium wilt of safflower (Carthamus tinctorius) caused by Fusarium oxysporum. Annual Report of Plant Protection of North Japan 59:90–93
Sharma M, Nagavardhini A, Thudi M, Ghosh R, Pande S, Varshney RK (2014) Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genomics 15:454
Shende SS, Wankhade DJ, Rajurkar AB, Varun P, Patil LD (2015) Characterization of pathogenic species of Fusarium oxysporum isolates of safflower by RAPD marker. Indian Journal of Plant Protection 43:251–253
Singh N, Kapoor R (2018) Quick and accurate detection of Fusarium oxysporum f. sp. carthami in host tissue and soil using conventional and real-time PCR assay. World Journal of Microbiology and Biotechnology 34:175
Singh AK, Chakrabarti DK, Chaudhary KB (1975) Two new diseases of safflower from India. Current Science 44:397–399
Singh V, Ranaware AM, Nimbkar N (2008) Breeding for Fusarium wilt resistance in safflower. In safflower: unexploited potential and world adaptability, proceedings of the 7th international safflower conference. Wagga Wagga, New South Wales, Australia
Smith JS, Chin EC, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theoretical and Applied Genetics 95:163–173
Sujatha M, Geetha AP, Sivakumar P, Palanisamy N (2008) Biotechnological interventions for genetic improvement of safflower. In: Proceedings of the 7th international safflower conference. Australian Oilseeds Federation. Wagga Wagga. pp. 3–6
Thangavelu R, Kumar KM, Devi PG, Mustaffa MM (2012) Genetic diversity of Fusarium oxysporum f. sp. cubense isolates (Foc) of India by inter simple sequence repeats (ISSR) analysis. Molecular Biotechnology 51:203–211
Velasco L, Fernandez-Martinez JM (2001) Breeding for oil quality in safflower. In: Bergman JW, Henning Mundel H (Eds.) Safflower - a multipurpose species with unexploited potential and world adaptability. Proceedings of the 5th international safflower conference. Williston. pp. 133–137
Wang B, Brubaker CL, Summerell BA, Thrall PH, Burdon JJ (2010) Local origin of two vegetative compatibility groups of Fusarium oxysporum f. sp. vasinfectum in Australia. Evolutionary Applications 3:505–524
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols - a guide to methods and applications. Academic Press, San Diego, pp 315–322
Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX (1999) POPGENE 3.2, user-friendly shareware for population genetic analysis. In: Molecular biology and biotechnology center. University of Alberta, Edmonton
Yuan L, Mi N, Liu S, Zhang H, Li Z (2013) Genetic diversity and structure of the Fusarium oxysporum f. sp. lini populations on linseed (Linum usitatissimum) in China. Phytoparasitica 41:391–401
Zayed MA, Yehia AH, El-Sebaey MA, Gowily AM (1980) Studies on the host-parasite relationship of safflower root rot disease caused by Fusarium oxysporum Schlecht. Egyptian Journal of Phytopathology 12:63–70
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
This work was funded by an R&D grant provided by the Research Council of University of Delhi, New Delhi, India. Neeraja Singh and Garima Anand are grateful to University Grants Commission, India for the research fellowships.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Denita H. Guerry
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure S1
Collection sites of Fusarium oxysporum f. sp. carthami isolates from safflower growing states of India. Numerals indicate the isolates collected from a particular district (a) Telangana (b) Karnataka (c) Maharashtra (d) Madhya Pradesh. (PNG 2790 kb)
Supplementary Figure S2
Disease reaction of safflower cultivar HUS-305, (a) resistant against isolate Foc 38; (b) moderately-susceptible against isolate Foc 9; and (c) susceptible against isolate Foc 1. (JPG 1360 kb)
Supplementary Figure S3
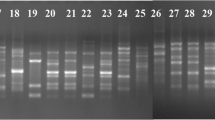
DNA fingerprinting profiles of 34 Fusarium oxysporum f. sp. carthami isolates obtained with (a) RAPD primer OPI-12 (b) ISSR primer ISSR-15. M, 100 bp DNA ladder (JPG 365 kb)
Supplementary Figure S4
Neighbor-joining dendrogram of Fusarium oxysporum f. sp. carthami (Foc) isolates based on 17 polymorphic RAPD markers. Three NJ clusters are designated as R1, R2 and R3. The column on the right of the dendrogram indicates the state from where the isolate was collected. (JPG 70 kb)
Supplementary Figure S5
Neighbor-joining dendrogram of Fusarium oxysporum f. sp. carthami (Foc) isolates based on 17 polymorphic ISSR markers. Two NJ clusters are designated as I1 and I2. The column on the right of the dendrogram indicates the state from where the isolate was collected. (JPG 73 kb)
Supplementary Figure S6
Population structure of Fusarium oxysporum f. sp. carthami isolates based on combined marker data. (a) Highest delta K value is observed for K value 2, indicating two sub-populations among the Foc isolates used in the study. (b) Each vertical bar represents a single Foc isolate. Admixture individuals have bars composed of two colors. (JPG 35 kb)
Supplementary Table 1
(DOCX 30 kb)
Rights and permissions
About this article
Cite this article
Singh, N., Anand, G. & Kapoor, R. Virulence and genetic diversity among Fusarium oxysporum f. sp. carthami isolates of India using multilocus RAPD and ISSR markers. Trop. plant pathol. 44, 409–422 (2019). https://doi.org/10.1007/s40858-019-00303-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-019-00303-1