Abstract
Purpose
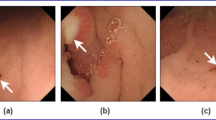
Wireless capsule endoscopy (WCE) is a fundamental diagnosing tool for gastro-intestinal (GI) lesion detection. Detecting and locating the lesions in WCE images using a computer-aided detection method is a challenging task because of the complex nature of GI systems and higher similarities between normal muscle and lesion regions. This study presents the lesion attention aware convolutional neural network (CNN) model using the self-attention mechanism to localize the lesion regions in the WCE image.
Method
The proposed novel lesion region estimator model uses ResNet-50 as a convolutional stem and self-attention mechanism that accurately aggregate spatial features in the global context to localize the lesion attention maps in WCE images. These lesion attention maps are fused with the original WCE image to elevate the lesion region in original WCE images. The lesion attention map estimator and classification network are mutually trained together to improve the detection accuracy of the lesion attention map estimator and the classification accuracy of WCE images respectively.
Results
Also, the model is tested on two publicly available datasets namely bleeding dataset and Kvasir-Capsule dataset with the overall classification accuracy of 95.1 and 94.7, respectively. The proposed attention augmented CNN model outperforms existing CNN-based models.
Conclusion
The experiment results show that the proposed lesion aware classification network offers superior classification accuracy thus aggregating semantic and conceptual attention maps using self-attention mechanisms. Further, this mechanism helps to improve the model explainability by analyzing the gradients of the attention maps.






Similar content being viewed by others
References
Soffer, S., Klang, E., Shimon, O., Nachmias, N., Eliakim, R., Ben-Horin, S., Kopylov, U., & Barash, Y. (2020). Deep learning for wireless capsule endoscopy: A systematic review and meta-analysis. Gastrointestinal Endoscopy, 92(4), 831–839. https://doi.org/10.1016/j.gie.2020.04.039
Ahn, J., Loc, H. N., Balan, R. K., Lee, Y., & Ko, J. (2018). Finding small-bowel lesions: Challenges in endoscopy-image-based learning systems. Computer, 51(5), 68–76. https://doi.org/10.1109/MC.2018.2381116
Guo, X., & Yuan, Y. (2020). Semi-supervised WCE image classification with adaptive aggregated attention. Medical Image Analysis, 64, 101733. https://doi.org/10.1016/j.media.2020.101733
Charfi, S., & El Ansari, M. (2020). A locally based feature descriptor for abnormalities detection. Soft Computing, 24(6), 4469–4481. https://doi.org/10.1007/s00500-019-04208-8
Ghosh, T., Fattah, S. A., & Wahid, K. A. (2018). CHOBS: Color histogram of block statistics for automatic bleeding detection in wireless capsule endoscopy video. IEEE Journal of Translational Engineering in Health and Medicine, 6, 1–12. https://doi.org/10.1109/JTEHM.2017.2756034
Gadermayr, M., Wimmer, G., Kogler, H., Vécsei, A., Merhof, D., & Uhl, A. (2018). Automated classification of celiac disease during upper endoscopy: Status quo and quo vadis. Computers in Biology and Medicine, 102, 221–226. https://doi.org/10.1016/j.compbiomed.2018.04.020
Yuan, Y., Yao, X., Han, J., Guo, L., & Meng, M. Q. H. (2017). Discriminative joint-feature topic model with dual constraints for WCE classification. IEEE Transactions on Cybernetics, 48(7), 2074–2085. https://doi.org/10.1109/TCYB.2017.2726818
Shen, Y., Guturu, P., & Buckles, B. P. (2011). Wireless capsule endoscopy video segmentation using an unsupervised learning approach based on probabilistic latent semantic analysis with scale invariant features. IEEE Transactions on Information Technology in Biomedicine, 16(1), 98–105. https://doi.org/10.1109/TITB.2011.2171977
Lan, L., Ye, C., Wang, C., & Zhou, S. (2019). Deep convolutional neural networks for WCE abnormality detection: CNN architecture, region proposal and transfer learning. IEEE Access, 7, 30017–30032. https://doi.org/10.1109/ACCESS.2019.2901568
Yu, J. S., Chen, J., Xiang, Z. Q., & Zou, Y. X. (2015). A hybrid convolutional neural networks with extreme learning machine for WCE image classification. In: 2015 IEEE International Conference on Robotics and Biomimetics (ROBIO). IEEE. pp. 1822–1827. DOI: https://doi.org/10.1109/ROBIO.2015.7419037.
Seguí, S., Drozdzal, M., Pascual, G., Radeva, P., Malagelada, C., Azpiroz, F., & Vitrià, J. (2016). Generic feature learning for wireless capsule endoscopy analysis. Computers in Biology and Medicine, 79, 163–172. https://doi.org/10.1016/j.compbiomed.2016.10.011
Gao, Y., Lu, W., Si, X., & Lan, Y. (2020). Deep model-based semi-supervised learning way for outlier detection in wireless capsule endoscopy images. IEEE Access, 8, 81621–81632. https://doi.org/10.1109/ACCESS.2020.2991115
Zhou, T., Han, G., Li, B. N., Lin, Z., Ciaccio, E. J., Green, P. H., & Qin, J. (2017). Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Computers in Biology and Medicine, 85, 1–6. https://doi.org/10.1016/j.compbiomed.2017.03.031
Chen, H., Wu, X., Tao, G., & Peng, Q. (2017). Automatic content understanding with cascaded spatial–temporal deep framework for capsule endoscopy videos. Neurocomputing, 229, 77–87. https://doi.org/10.1016/j.neucom.2016.06.077
Yuan, Y., & Meng, M. Q. H. (2017). Deep learning for polyp recognition in wireless capsule endoscopy images. Medical physics, 44(4), 1379–1389. https://doi.org/10.1002/mp.12147
Sekuboyina, A. K., Devarakonda, S. T., & Seelamantula, C. S. (2017). A convolutional neural network approach for abnormality detection in wireless capsule endoscopy. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE. pp. 1057–1060. DOI: https://doi.org/10.1109/ISBI.2017.7950698.
Iakovidis, D. K., Georgakopoulos, S. V., Vasilakakis, M., Koulaouzidis, A., & Plagianakos, V. P. (2018). Detecting and locating gastrointestinal anomalies using deep learning and iterative cluster unification. IEEE Transactions on Medical Imaging, 37(10), 2196–2210. https://doi.org/10.1109/TMI.2018.2837002
Aoki, T., Yamada, A., Aoyama, K., Saito, H., Tsuboi, A., Nakada, A., Niikura, R., Fujishiro, M., Oka, S., Ishihara, S., Matsuda, T., & Tada, T. (2019). Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointestinal Endoscopy, 89(2), 357–363. https://doi.org/10.1016/j.gie.2018.10.027
Gomes, S., Valério, M. T., Salgado, M., Oliveira, H. P., & Cunha, A. (2019). Unsupervised neural network for homography estimation in capsule endoscopy frames. Procedia Computer Science, 164, 602–609. https://doi.org/10.1016/j.procs.2019.12.226
Wang, S., Xing, Y., Zhang, L., Gao, H., & Zhang, H. (2019). Deep convolutional neural network for ulcer recognition in wireless capsule endoscopy: Experimental feasibility and optimization. Computational and Mathematical Methods in Medicine. https://doi.org/10.1155/2019/7546215
Alaskar, H., Hussain, A., Al-Aseem, N., Liatsis, P., & Al-Jumeily, D. (2019). Application of convolutional neural networks for automated ulcer detection in wireless capsule endoscopy images. Sensors, 19(6), 1265. https://doi.org/10.3390/s19061265
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., Kaiser, Ł, & Polosukhin, I. (2017). Attention is all you need. Advances in Neural Information Processing Systems, 2017, 5998–6008.
Muruganantham, P., & Balakrishnan, S. M. (2021). A survey on deep learning models for wireless capsule endoscopy image analysis. International Journal of Cognitive Computing in Engineering, 2(February), 83–92. https://doi.org/10.1016/j.ijcce.2021.04.002
Münzer, B., Schoeffmann, K., & Böszörmenyi, L. (2018). Content-based processing and analysis of endoscopic images and videos: A survey. Multimedia Tools and Applications, 77(1), 1323–1362. https://doi.org/10.1007/s11042-016-4219-z
Rathnamala, S., & Jenicka, S. (2021). Automated bleeding detection in wireless capsule endoscopy images based on color feature extraction from Gaussian mixture model superpixels. Medical & Biological Engineering & Computing, 59(4), 969–987. https://doi.org/10.1007/s11517-021-02352-8
Coimbra, M. T., & Cunha, J. S. (2006). MPEG-7 visual descriptors—contributions for automated feature extraction in capsule endoscopy. IEEE Transactions on Circuits and Systems for Video Technology, 16(5), 628–637.
Karargyris, A., & Bourbakis, N. (2011). Detection of small bowel polyps and ulcers in wireless capsule endoscopy videos. IEEE Transactions on Biomedical Engineering, 58(10), 2777–2786. https://doi.org/10.1109/TBME.2011.2155064
Li, B., & Meng, M. Q. H. (2012). Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Transactions on Information Technology in Biomedicine, 16(3), 323–329. https://doi.org/10.1109/TITB.2012.2185807
Yuan, Y., Li, B., & Meng, M. Q. H. (2015). Improved bag of feature for automatic polyp detection in wireless capsule endoscopy images. IEEE Transactions on Automation Science and Engineering, 13(2), 529–535. https://doi.org/10.1109/TASE.2015.2395429
Yuan, Y., Li, B., & Meng, M. Q. H. (2016). WCE abnormality detection based on saliency and adaptive locality-constrained linear coding. IEEE Transactions on Automation Science and Engineering, 14(1), 149–159. https://doi.org/10.1109/TASE.2016.2610579
Khan, M. A., Rashid, M., Sharif, M., Javed, K., & Akram, T. (2019). Classification of gastrointestinal diseases of stomach from WCE using improved saliency-based method and discriminant features selection. Multimedia Tools and Applications, 78(19), 27743–27770. https://doi.org/10.1007/s11042-019-07875-9
Al Mamun, A., Hossain, M. S., Hossain, M. M., & Hasan, M. G. (2019). Discretion way for bleeding detection in wireless capsule endoscopy images. In: 2019 1st International Conference on Advances in Science, Engineering and Robotics Technology (ICASERT). IEEE. pp. 1–6. DOI: https://doi.org/10.1109/ICASERT.2019.8934589.
Ghosh, T., & Chakareski, J. (2021). Deep transfer learning for automated intestinal bleeding detection in capsule endoscopy imaging. Journal of Digital Imaging. https://doi.org/10.1007/s10278-021-00428-3
Ali, H., Sharif, M., Yasmin, M., Rehmani, M. H., & Riaz, F. (2020). A survey of feature extraction and fusion of deep learning for detection of abnormalities in video endoscopy of gastrointestinal-tract. Artificial Intelligence Review, 53(4), 2635–2707. https://doi.org/10.1007/s10462-019-09743-2
Li, M., Hsu, W., Xie, X., Cong, J., & Gao, W. (2020). SACNN: Self-attention convolutional neural network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Transactions on Medical Imaging, 39(7), 2289–2301.
Xing, X., Yuan, Y., & Meng, M. Q. H. (2020). Zoom in lesions for better diagnosis: Attention guided deformation network for WCE image classification. IEEE Transactions on Medical Imaging, 39(12), 4047–4059. https://doi.org/10.1109/TMI.2020.3010102
Hu, J., Shen, L., & Sun, G. (2018). Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 7132–7141
Woo, S., Park, J., Lee, J. Y., &Kweon, I. S. (2018). Cbam: Convolutional block attention module. In: Proceedings of the European Conference on Computer Vision (ECCV). pp. 3–19.
Bello, I., Zoph, B., Vaswani, A., Shlens, J., & Le, Q. V. (2019). Attention augmented convolutional networks. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (pp. 3286–3295)
Wang, Z., Zou, N., Shen, D., & Ji, S. (2020). Non-local u-nets for biomedical image segmentation. Proceedings of the AAAI Conference on Artificial Intelligence., 34(04), 6315–6322.
Khanh, T. L. B., Dao, D. P., Ho, N. H., Yang, H. J., Baek, E. T., Lee, G., Kim, S. H., & Yoo, S. B. (2020). Enhancing u-net with spatial-channel attention gate for abnormal tissue segmentation in medical imaging. Applied Sciences, 10(17), 5729. https://doi.org/10.3390/app10175729
Huang, G., Zhu, J., Li, J., Wang, Z., Cheng, L., Liu, L., & Zhou, J. (2020). Channel-attention U-Net: Channel attention mechanism for semantic segmentation of esophagus and esophageal cancer. IEEE Access, 8, 122798–122810. https://doi.org/10.1109/ACCESS.2020.3007719
Ren, X., Huo, J., Xuan, K., Wei, D., Zhang, L., & Wang, Q. (2020). Robust brain magnetic resonance image segmentation for hydrocephalus patients: Hard and soft attention. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) (pp. 385–389). IEEE. DOI: https://doi.org/10.1109/ISBI45749.2020.9098541.
Chen, B., Li, J., Lu, G., & Zhang, D. (2019). Lesion location attention guided network for multi-label thoracic disease classification in chest X-rays. IEEE Journal of Biomedical and Health Informatics, 24(7), 2016–2027. https://doi.org/10.1109/JBHI.2019.2952597
Fong, C. (2014). Analytical methods for squaring the disc. Retrieved from https://arxiv.org/abs/1509.06344. pp. 1–33
Deeba, F., Islam, M., Bui, F. M., & Wahid, K. A. (2018). Performance assessment of a bleeding detection algorithm for endoscopic video based on classifier fusion method and exhaustive feature selection. Biomedical Signal Processing and Control, 40, 415–424. https://doi.org/10.1016/j.bspc.2017.10.011
Smedsrud, P. H., Thambawita, V., Hicks, S. A., Gjestang, H., Nedrejord, O. O., Næss, E., Borgli, H., Jha, D., Berstad, T. J. D., Eskeland, S. L., Lux, M., & Halvorsen, P. (2021). Kvasir-Capsule, a video capsule endoscopy dataset. Scientific Data, 8(1), 1–10. https://doi.org/10.1038/s41597-021-00920-z
Pogorelov, K., Ostroukhova, O., Petlund, A., Halvorsen, P., de Lange, T., Espeland, H. N., Kupka, T., Griwodz, C., Riegler, M. (2018). Deep learning and handcrafted feature based approaches for automatic detection of angiectasia. In: 2018 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI). IEEE. pp. 365–368. DOI: https://doi.org/10.1109/CBMS.2018.00073.
Xing, X., Yuan, Y., Jia, X., & Meng, M. Q. H. (2019). A saliency-aware hybrid dense network for bleeding detection in wireless capsule endoscopy images. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE. pp. 104–107
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Muruganantham, P., Balakrishnan, S.M. Attention Aware Deep Learning Model for Wireless Capsule Endoscopy Lesion Classification and Localization. J. Med. Biol. Eng. 42, 157–168 (2022). https://doi.org/10.1007/s40846-022-00686-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-022-00686-8