Abstract
The augmented randomized complete block design (ARCBD) is widely used in plant breeding programs to screen numerous new treatments. The error variance is estimated based on the replicated control treatments run over a randomized complete block design and is used to test the new treatments that are administrated each once in the extended units of the blocks. However, one or more observations corresponding to the control treatments may be missed in experiments, making difficulties, e.g., biased estimates. An approximate common approach to deal with this problem is the imputation of the estimated value which is with some uncertainties. Moreover, in real-life experiments, there are more sources of uncertainty that cause conflict-indeterminate, vague, imprecise, and erroneous data that increases the complexity of the analysis. In this paper, an exact scheme is utilized to deal with a missing control treatment in ARCBD. To overcome the problem of indeterminacies in data, a novel neutrosophic analysis approach is proposed. Specifically, the problem of a missing value in an ARCBD for an uncertain environment is resolved analytically by considering an augmented incomplete block design in the framework of neutrosophic statistics so-called neutrosophic augmented randomized complete block design (NARCBD). In this approach, by proposing the neutrosophic model, the neutrosophic estimations as well as the mathematical neutrosophic adjusted sums of squares are derived and the analysis of variance table is provided. The new model is applied to the neutrosophic genotype data example of safflower and assessed by a simulation study. Furthermore, a code in the R software was written to analyze the data based on the proposed approach to fill the calculation gap for data analysis in NARCBD with a missing value. In light of the results observed, it can be concluded that the neutrosophic exact proposed method performs better than the classic in the presence of uncertainty.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In an experiment, missing observations are often arisen, particularly when the experimental units are breeding or biological varieties. Consider, for instance, a long-term experiment in which different safflower genotypes were planted in a blocking design to determine their yields. During the experiment, it is very likely that one or more plants of a variety in a block be missed due to a limited experimental unit or being withered; this will result in the problem of missing an observation completely at random. The missing values could cause a major problem in the statistical analysis of an experiment and might adversely affect the results, causing bias in treatment effect estimation and the upward sum of squares, increasing the estimate variability, or may diminish the power of testing treatment effects. Therefore, the missing problem should be carefully treated. The attempts to resolve such a problem go back many years ago when it was suggested that the vacant cell be filled by the estimation of the lost observation [1, 2]. Later, the missing problem has been studied for different designs; Cornish [3] discussed how to estimate missing values in incomplete block designs (IBD). Baird and Kramer [4] considered the Balanced IBD with several missing values. Apart from the pre-mentioned estimating method of missing observations, an exact method to deal with the missing values has been considered by some researchers, which is the so-called “general regression significance test” [5]. This method has been mostly studied for the analysis of covariance [6,7,8]. Recently, the exact scheme is highlighted by Sirikasemsuk, et al. [9] where the adjusted regression sum of squares of the randomized complete block design (RCBD) with a missing observation was calculated mathematically.
The problem of missing values is less discussed in ARCBD. ARCBD is an unbalanced design that is obtained from an RCBD, including the control treatments, by distributing a numerous number of added new genotypes randomly to blocks with the aim of studying them. ARCBD has been introduced by Federer [10] to deal with the screening stage of the breeding program, where the experimenters face a large number of new treatments (genotypes) (see also [11] for more details). Consider a randomized complete block design where genotypes are augmented randomly in all of the blocks by proliferating the number of experimental units within blocks. Therefore, each block includes only once each of the c control treatments, so-called checks. Therefore, the blocks and treatment effects are orthogonal. The genotypes run without replication in the experiment; that is, each appears only in one of the blocks. Several classes of Augmented block designs have been developed. Federer and Crossa [11] give a review of augmented designs. [12,13,14] developed SAS code for analyzing the data that come from augmented designs.
As mentioned above, a common approach is to estimate the missing value and used it for analyzing the data. This is a method so-called approximate [5]; however, the estimated value intrinsically differs from the original value and hence may cause a bias in results. There are two issues with this approach. First is the uncertainty about the estimated missing value especially, if there are some ambiguities and indeterminacies in the data itself and the second is the potential for the inefficiency of the approximate approach. Neither the classic analysis is able to provide information regarding the measure of indeterminacy to analyze the genotypes precisely nor the approximate method is enough reliable to make inferences, unbiasedly.
In classic statistics, all classes of augmented designs have been considered and analyzed, yet to the best of our knowledge, there is still a vacancy for these designs in the uncertain environment. Accordingly, several researchers have employed fuzzy sets and intuitionistic fuzzy sets in their investigations to effectively tackle the presence of uncertainty and ambiguity within the data. In this context, Kumar [15] highlighted a new method for addressing the type-2 intuitionistic fuzzy transportation problem. Naveed et al. [16] have focused on the analysis of interval-valued fuzzy soft sets and the application of IVFSS algorithm in the risk assessment of prostate cancer. Kumar [17] provided an innovative approach to address the challenges posed by intuitionistic fuzzy solid transportation problems. Kumar [18] furnished a facile approach toward resolving type-2 and type-4 fuzzy transportation predicaments. Kumar [19] introduced a novel technique for resolving transportation problems that fall under the type-1 and type-3 fuzzy categories. A novel category of orthopair fuzzy sets called “ (2, 1) -fuzzy sets” was proposed by Al-shami [20]. These sets employ a pair of values within the unit interval to indicate the degrees of membership and non-membership. A comparison with other fuzzy set types was made, and essential operations and properties were suggested alongside their potential applications in multi-criteria decision-making techniques. Al-shami and Mhemdi [21] presented a generalized frame for orthopair fuzzy sets known as (m, n) -fuzzy sets, along with their practical applications in methods for making decisions involving multiple criteria. Al-shami et al. [22] introduced a novel extension of fuzzy soft sets known as (a, b) -fuzzy soft sets.
Although fuzzy sets and intuitionistic fuzzy sets can be viewed as a broader space of classical set theory, neutrosophic sets surpass fuzzy sets by introducing a third parameter called indeterminacy. Indeterminacy assesses the degree of uncertainty or incompleteness in a statement, improving the representation of uncertainty by incorporating not only the degree of membership and non-membership but also the degree of indeterminacy. Besides, computations with neutrosophic sets are less complex, leading to more efficient use of time and resources. Thus, neutrosophic sets are more effective than intuitionistic fuzzy sets in clustering complex datasets with overlapping information and can manage multiple degrees of overlap and contradictions that intuitionistic fuzzy sets struggle with. There are more discussions in the literature about the differences between neutrosophic statistics (NS) and other types of statistics. Smarandache [23] demonstrated that NS are more general than interval statistics in many cases and gave examples of how neutrosophic algebra can provide greater accuracy. For instance, NS deals with all types of indeterminacy, whereas interval statistics only deals with indeterminacy represented by intervals. Accordingly, neutrosophy has led to the development of new mathematical theories, such as neutrosophic set theory, probability, statistics, and logic. These theories generalize classical and fuzzy counterparts and have practical applications in various fields.
We have considered the missing value in ARCBD and raised some ambiguities in data, such as the indeterminacy, the pre-unknown number of new treatments in ARCBD, erroneous estimating of the missing value, and imprecise F test. The probable inadequacy of the classic and fuzzy methods to deal with such complexity motivated us to utilize NS. In this paper, the indeterminacies are tackled by examining the analysis of missing values in NS framework, which provides additional information about the level of uncertainty. Moreover, an exact method for dealing with a missing value was suggested through the NS to came up with a more precise analysis in ARCBD. This is for the first time that the augmented block design is discussed in neutrosophic environment, while also focusing on the issue of missing value.
The concept of NS has been introduced by Smarandache [24]. Aslam [25] explained the differences between fuzzy statistics, NS, and classic statistics. As Smarandache [23] explained, NS is much broader than interval statistics because it deals with all types of indeterminacy including non-well-known sample size, neutrosophic random variable, and neutrosophic probability distributions. Aslam [26] has discussed the neutrosophic ANOVA method. AlAita and Aslam [27] highlighted the application of neutrosophic analysis of covariance to neutrosophic completely random designs, neutrosophic randomized complete block designs, and neutrosophic split-plot designs. The utilization of neutrosophic statistical analysis for split-plot designs was discussed by AlAita et al. [28]. [29] suggested post hoc multiple comparison tests under NS. Neutrosophic correlation and simple linear regression have been discussed by Salama et al. [30]. Analysis of neutrosophic multiple regression has been suggested by Nagarajan, et al. [31]. Jafar et al. [32] proposed a novel approach for site selection in solid waste management systems through the development of distance and similarity measures using max–min operators of neutrosophic hypersoft sets. Jafar et al. [33] have constructed trigonometric similarity metrics for neutrosophic hypersoft sets and employed them to select renewable energy sources. Jafar et al. [34] proposed utilizing similarity measures such as tangent, cotangent, and cosine within a neutrosophic environment for the purpose of selecting academic programs. The matrix theory for neutrosophic hypersoft set and its application in multiattributive multicriteria decision-making problems were analyzed by Jafar and Saeed [35]. In the very most recent years, numerous neutrosophic statistical studies have been discussed [36,37,38,39,40,41,42,43,44].
Neutrosophic normality
The following offers some basic concepts regarding the neutrosophic random variables that will be advantageous in the subsequent sections.
The \(X_N{\in }\left[ X_L{,\ }X_U\right] \) is a neutrosophic random variable with indeterminacy interval, \(I_N,\) and is written as \(X_N{=\ }X_L{+\ }X_UI_N\), where \(X_L\) is determinate part and \(X_UI_N\) is indeterminate part, where \(I_N{\in }{[}I_L,I_U{]}\) is measure of uncertainty. Clearly, \(X_N\) is reduced to the classical random variable at \(I_L=0.\)
Definition 1
[28] Suppose that \(X_N\) is a neutrosophic random variable with neutrosophic normal distribution with population mean and variance \({\mu }_N\) and \({\sigma }^{{2}}_N,\) respectively if the neutrosophic mean and variance can be found as
Definition 2
[28] Suppose a random sample of size n, selected from a population of size N, contains indeterminate observations. The estimated neutrosophic sample mean \({{\overline{x}}}_N\) and the variance \(s^{{2}}_N\), are expressed by;
Neutrosophic model of augmented randomized complete and incomplete block designs
The neutrosophic statistical model for a NARCBD with a blocks, c checks and v new treatments can be formulated as follows:
The neutrosophic form of \(y_{Nhijg}\) can be expressed as
where \(h=l\ or\ q\) stands for the neutrosophic effects associated with new treatments or checks, respectively, \({\mu }_N\) is a neutrosophic overall mean, \({\alpha }_{Ni}\) is the neutrosophic effect of the ith block, \({\tau }_{Nqj}\) is the neutrosophic effect of the jth check, \({\tau }_{Nlig}\) is the neutrosophic effect of the g th new treatment in the ith block, and \({\varepsilon }_{Nhijg}\) is the random error with neutrosophic zero mean and variance \({\sigma }^2_N\). It is assumed that the errors are independent and neutrosophic normally distributed. Let \({v}=\sum ^b_{i=1}{n_{(li)}}\) refer to the number of new treatments, c for the number of check treatments, and b for the number of blocks; therefore,\(e={v}+c\) is the total number of new and check treatments and the total number of all plots in the blocks is n; i.e., \(n={v}+bc\). Formula (1) is also the model for neutrosophic augmented incomplete block design (NAIBD) in which one or more c checks do not appear in some blocks; therefore, \(n<{v}+bc.\) Throughout the paper in the context of neutrosophic ANOVA, the \({SS}_{NT},\ {SS}_{NTr},\ {SS}_{NB}\) and \({SS}_{NE}\), \({SS}_{N.}{\in }\left[ {SS}_{L.},\ {SS}_{U.}\right] ,\) stand for the neutrosophic sum of squares (\( SS_N \)) total, treatment, block, and error, respectively, and the subscript N denotes the neutrosophic context.
Estimation of neutrosophic parameters
Neutrosophic approximate analysis (NAA)
The neutrosophic analysis for an ARCBD is studied by AlAita and Talebi [45]. Now, consider the problem of losing a check in a block completely at random; for example, rth check in block m. In the classic statistics, the missing observation is estimated by minimizing the sum of squares errors and assumed as the true value to analyze the data by lessening one degrees of freedom of the error. Such an approach can be extended to NS by estimating the neutrosophic missing observation by \(y^*_{Nqmr}=\frac{{{by}_{Nqm.}+cy}_{Nq.r}-y_{Nq..}}{\left( c-1\right) \left( b-1\right) }=[y^*_{Lqmr},\ y^*_{Uqmr}]\), where throughout the paper the notion (.) means the sum over the related indices. However, a poor estimate of missing observation may lead to inefficient inference about treatments. A simulation study will be subsequently conducted for a more detailed analysis of this issue.
Neutrosophic exact analysis (NEA)
For a more precise analysis of data that includes missing values in the NARCBD, an exact analytical approach is proposed to address neutrosophic missing values in the NARCBD. By missing a check in NARCBD, one face with an incomplete block hence the design is a NAIBD in terms of control treatments, regardless of the new treatments. The design that involves a missing check treatment within a block is regarded as an NAIBD and will be analyzed, accordingly. Note that the unbalanced blocks deface the orthogonality between blocks and checks due to not having all checks in each block. Consequently, this contravenes control and new treatments estimation and gives invalid sums of squares which are not sum to a right total. To response to the need of the required adjustment, the exact approach is accomplished based on the complete case analysis of the observations for the NAIBD. It should be noted that the results in NAIBD is a generalization of NIBD where the new treatments will be ignored.
To estimate the model parameters of a NAIBD where it is assumed an observation is missing completely at random, e.g., the rth check treatment in the mth block for any r and m, the least squares normal equations (NEs) are
By solving NEs (2–5) under the constraints \(\sum ^b_{i{=1}}{{{\hat{\alpha }}}_{Ni}{=0}}\), \(\sum ^c_{j{=1}}{{{\hat{\tau }}}_{Nqj}}+\sum ^b_{i{=1}}{\sum ^{n_{li}}_{g{=1}}{{{\hat{\tau }}}_{Nlig}{=0}}}\), the neutrosophic estimates of the parameters in (1) are
where \({{\hat{\mu }}}_{Nq}=\frac{{(}bc{-}b{-}c{)}y_{Nq..}+by_{Nqm.}+cy_{Nq.r}}{bc{(}b{-}{1)(}c{-1)}}.\)
In the same manner, as above, the neutrosophic estimates of the parameters of the reduced treatment- and block-models, obtained by deleting the block and treatment effects from (1), can be calculated, respectively.
Neutrosophic testing of effects
To test the neutrosophic parameters in NARCBD, a need is to calculate the \({SS}_{NT}\) and neutrosophic adjusted(adj) and unadjusted(unadj) sums of squares for the blocks, treatments (new and check) and the NSS for error in the NARCBD.
neutrosophic mean squares and neutrosophic test statistics \(F_N\) are obtained as in the NANOVA Tables 1 and 2. The \(F_N\in [F_L,F_U]\), and the neutrosophic form of \(F_N\) is \(F_N=F_L+F_UI_{F_N};\ I_{F_N}\in [I_{F_L},I_{F_U}]\), where \(F_L\) and \(F_UI_{F_N}\) are determinate and indeterminate parts of each proposed test (see Aslam [46]). This test reduces to a test under classic statistic if \(I_{F_N}\) = 0 and \(I_{F_N}\)can be calculated by \({\left( F_U-F_L\right) }/{F_U.}\) Accordingly, the neutrosophic p value is \(p_N\in \left[ p_L,p_U\right] ,\) where \(p_L\) and \(p_U\) correspond to \(F_L\) and \(F_U\), respectively.
Neutrosophic hypotheses and decision rules
To test the blocks, checks and new treatments the null and alternative hypotheses are:
\( H_{N0}:{\alpha }_{Ni}=0\ \ {v}s\ \ H_{N1}:\mathrm{at\ least\ one\ \alpha }_{Ni}\ne 0,~i=1,\ 2,\ldots ,\ b, \)
\( H_{N0}:{\tau }_{Nqj}=0\ \ {v}s\ \ H_{N1}:\mathrm{at\ least\ one\ \tau }_{Nqj}\ne 0,~j=1,\ 2,\ldots ,\ c, \)
\( H_{N0}:{\tau }_{Nlig}=0\ \ {v}s\ \ H_{N1}:\mathrm{at\ least\ one\ \tau }_{Nlig}\ne 0,~g=1,\ 2,\ldots ,\ n_{(li)}. \)
The null hypothesis is not rejected if \(\textrm{min}\{p_\mathrm{N-value}\}\ge \alpha \), where \(\alpha \) is a level of significance. Conversely, the null hypothesis is rejected if \(\textrm{max}\left\{ p_\mathrm{N-value}\right\} <\alpha \). Under the neutrosophic normality assumption and if \(H_{N0}\) is correct, the \(p_N\)-value is obtained from F-distribution with the related degrees of freedom as given in the ANOVA Tables. Following [10], we present two ANOVA tables for unadjusted (A) and adjusted blocks (B), respectively. Two ANOVA tables are provided, one for unadjusted blocks (A) and the other for adjusted blocks (B). In the NANOVA Tables 1 and 2, the calculation formulas for testing the formulated neutrosophic hypotheses have been summarized for a NIBD as a NARCBD with a missing control treatment. In Table 2, the New by check interaction effect with one degree of freedom is separated which allows for testing the new genotypes. We provide a code in R for computing these NANOVA Tables. A code in R is made available for the computation of these NANOVA Tables.
Numerical example and simulation
In this section, the proposed NS for dealing with a missing value in an ARCBD is numerically assessed. For this purpose, a set of real neutrosophic data with uncertain observations from NARCBD with a missing check will be analyzed. A simulation study will also be contrived for evaluation. To assess the efficiency of the neutrosophic exact method for missing value analysis, the \(F_N{\in }[F_L,F_U]\) tests for the treatment effects are calculated and compared with the existing tests under classic statistics in terms of uncertainty.
Real data example
An experiment was run in three parts of a farm at the Isfahan University of Technology to study the safflower plant breeding program. In this experiment, 99 cultivars of safflower (an oil and medicinal plant) genotypes were planted using an NARCBD with three blocks. Each block contains 8 control (check) treatments and 25 genotypes (new treatments). The oil seed functions of the plants were measured and recorded in Table 8 (see the Appendix). To assess the exact analysis approach in this study, a check treatment is deleted (for instance, G35 being eliminated from the third block). The NANOVA Tables for this set of data including sums of squares (\({SS}_N\)) and the observed F-test statistics (\(F_N\)) with corresponding p values (\(p_N\)) are calculated based on the neutrosophic approximate and the proposed exact methods of dealing with a missing value in NARCBD. The results for the approximate method are summarized in Table 3 based on the neutrosophic estimate of missing observation \(y^{{*}}_{Nqmr}=\frac{{{by}_{Nqm.}+cy}_{Nq.r}{-}y_{Nq..}}{\left( c{-1}\right) \left( b{-1}\right) }=\left[ {600.6071,}\ {602.9157}\right] .\) Neutrosophic ANOVA Tables 4 and 5 present the results of the exact method for dealing with the same missing observation.
Simulation study
Now, by the following simulation study, the efficiency of the exact approach will be compared to the approximate method, as well as classic and neutrosophic statistics in analyzing data that contains missing values in NARCBD. For this purpose, the empirical neutrosophic type I error and the power of the test for neutrosophic treatment effects at a given nominal significant level were calculated. The simulation was carried out for the given number of treatments and blocks which are already presented in various existing examples (e.g., [10]). The data are first generated from the corresponding neutrosophic normal distribution. Then, by assuming nominal significant levels at 0.05 and 0.01, the MC method was employed to compute both the empirical neutrosophic type I error and the power of the test for neutrosophic treatment effects. The simulation was replicated 10,000 times for both methods.
To calculate the neutrosophic empirical Type I error rate and the test power for an MC experiment, the following steps need to be completed (Figs. 1, 2):
MC simulation to compute \({{\alpha }}_{{\textrm{Empirical}}}\)
Step 1: The random sample \(x^{(i)}_{N1},x^{(i)}_{N2},\ldots ,\ x^{(i)}_{Nn}\) is generated from the neutrosophic normal standard distribution under \(H_{N0}\), \(i=1,2,\ldots ,10{,}000\).
Step 2: The \(F_{Ni}\)-test is computed under \(H_{N0}\).
Step 3: The results are recorded by assigning a value of \(I_{Ni}=1\) when the \(H_{N0}\) is rejected, and \(I_{Ni}=0\) otherwise.
Step 4: The ratio \(\frac{1}{10,000}\sum ^{10,000}_{i=1}{I_{Ni}}\) is computed and take it as \({\alpha }_{\textrm{Empirical}}\).
Step 5: The neutrosophic coefficient factor \(NCF=\frac{\alpha }{{\alpha }_{\textrm{Empirical}}}\) is computed.
MC simulation to compute \({\textrm{Power}}_{\textrm{Empirical}}\)
Step 1: The random sample \(x^{(i)}_{N1},~x^{(i)}_{N2},\dots ,\ x^{(i)}_{Nn}\) is generated from the neutrosophic normal standard distribution under \(H_{N1}\), \(i=1,2,\dots ,10{,}000\). For instance, \(({\mu }_{N1},{\mu }_{N2},{\mu }_{N3},{\mu }_{N4})=(0,1,2,4)\).
Step 2: The \(F_{Ni}\)-test is computed under \(H_{N1}\).
Step 3: The results are recorded by assigning a value of \(I_{Ni}=1\) when the \(H_{N1}\) is rejected, and \(I_{Ni}=0\) otherwise.
Step 4: The ratio \(\frac{1}{10,000}\sum ^{10,000}_{i=1}{I_{Ni}}\) is computed and take it as \(\textrm{Power}_\textrm{Empirical}\).
Step 5: The \(\textrm{Power}_\mathrm{Empirical({adj})}=NCF\times \textrm{Power}_\textrm{Empirical}\) is computed.
In this section, the calculations were completed using the R software. To achieve this, a code was created within the R program. The R program is a popular option for carrying out data analysis due to its strong statistical features and visualization capabilities. As an open-source programming language, users can access and customize the source code according to their specific requirements. Additionally, R offers a cost-effective solution for data analysis since it can be downloaded and used free of charge. R is frequently employed by data analysts and scientists, and many possess familiarity with its functions. The use of R can make the data science process unique and exciting because of its ability to perform various advanced and complex statistical computations. Moreover, R can present interpreted data graphically, making scientific reports easier to comprehend. The program’s capacity to compute enormous amounts of data quickly is also noteworthy [47,48,49,50]. Specifically, an R code was developed for this simulation study, taking advantage of the software’s exceptional ability to calculate and provide the necessary simulation materials.
The simulated neutrosophic type I error for exact and approximate methods were calculated and summarized in Table 6. The performance of the exact scheme is reliable due to having a close empirical type one error with the nominal \(\alpha \), but the approximate method exposes an inflated empirical type I error which means the null hypothesis is overly rejected. That is, the treatments may erroneously be considered significant. Meanwhile, the neutrosophic empirical type one error for both approximate and exact methods is better than the classic statistics. The superiority of exact over approximate method as well as neutrosophic over classic is evidently figured in Figs. 3 and 4. Figures 3 and 4 display classic empirical type I error, \({\alpha }_L\), and neutrosophic, \({\alpha }_N\), respectively, obtained for 200 times repetitions of the simulation. The empirical \({\alpha }_N\) for the exact method disperses around the nominal \(\alpha \)- level.
To measure the deviation proportion of empirical type I error from nominal more precisely, the neutrosophic coefficient factor (NCF) was defined as NCF = nominal \(\alpha {-}\textrm{level}/\textrm{Empirical}\) type I error = [ LCF, UCF] and calculated in the last columns of Table 6 for both approximate and exact methods. The closer NCF is to 1 prefigures the more precision of a method.
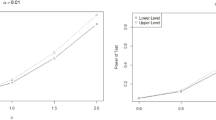
For more investigation, the power of testing the neutrosophic treatment effects using NANOVA was also calculated for both methods. The results are recorded in Table 7 for NARCBD with \(\left( b{,\ }c{,\ v,\ }n\right) =\left( {3,\ 4,\ 8,\ 20}\right) \) and different sets of check means, \(\left( {\mu }_{N1},~{\mu }_{N2},~{\mu }_{N3},~{\mu }_{N4}\right) \) and new treatments given in the Tables for 0.05 and 0.01 significant levels. For more understanding, the results in Table 7 are graphically displayed in Figures 5 and 6; without loss of generality, the powers were plotted in ascending order. Figure 5 demonstrates the power of the treatment test for the classic exact (upper bound) and approximate (lower bound) methods, revealing the superiority of the exact proposed method. Figure 6 is the neutrosophic version of Fig. 5 with the same conclusion for the superiority of the exact method. Moreover, the neutrosophic color spectrum above the classic lower bound indicates the superiority of the neutrosophic model.
Discussion
This study was administrated with the aim of improving the analysis of uncertain and indeterminate data in the framework of NS for an augmented block design with a missing value. In classic statistics, the unrecorded observation is estimated and is used as the true value to analyze the data; however, the estimated value is intrinsically neutrosophic due to not matching the original value, exactly. Therefore, it is logical to use NS to deal with the estimated missing data.
In this study, the basic analytical needs were formulated for ANOVA of an ARCBD in the neutrosophic context in the presence of a missing value, though it is also useable for classic statistics; then, the proposed method was assessed by a set of real safflower plant data, numerically. A simulation study was also performed to see how precise are the neutrosophic approximate and exact methods. To assess the performances of these methods the empirical type I error and the power of the test for treatment effects have been used. The neutrosophic results disclosed a better performance than the classic; moreover, the proposed exact method performance was more efficient than the approximate method. For more illustration, the results showed that the empirical \( \alpha \) is a little inflated than that of nominal level in the approximate method, while, it was compatible with the nominal \( \alpha \) for the exact method. Interestingly, the exact method, either classic or neutrosophic, indicated a higher power of test than the neutrosophic approximate method. The results in Tables 6 and 7 and Figs. 3, 4, 5, 6 reveal the superiority of the proposed approach in this study. More advantages of the proposed neutrosophic model are tabulated below. Some disadvantages are also listed.
The code written in the R program for NACRBD with missing values, given online in the supplemental material, provides the NANOVA in which the adjusted unbiased \( SS_N \) of neutrosophic treatment effect is directly calculated. Moreover, the neutrosophic treatment effect is decomposed to check and new treatments. This eventuated to having the \( F_N \)-test statistics and \( p_N \)-values for each effect, separately. Furthermore, the \( F_N \)-tests in NANOVA offer greater flexibility, applicability, and information when compared with the existing F-tests for classic ARCBD in uncertain environments. Moreover, the post hoc test for each of the neutrosophic treatments as well as the new by check interaction is presented in this program which is crucial for determining the significant new genotypes. This code fills the calculation gap and the mistaken analysis of the data in classic ACRBD with a missing value.
Advantages | Disadvantages |
|---|---|
The neutrosophic exact proposes approach in this study is more efficient than that of existing approximate method for dealing with missing value | Neutrosophic statistical models is more complex than traditional statistical models due to the incorporation of neutrosophic set theory |
Neutrosophic logic furnishes a mathematical structure for depicting levels of indeterminacy which can aid researchers in addressing the intricacies that arise from real-world data | The results obtained from neutrosophic statistical models may be difficult to be interpreted due to the inherent uncertainty in the data and model assumptions |
The neutrosophic model is more flexible and versatile that allows for the handling of uncertainty and indeterminacy in data and analysis in diverse contexts | Understanding of the indeterminacy in a system is both in a subjective and an objective sense such as uncertainty or vagueness that sometimes, it is hard to be redefined from a formal point of view |
Using a neutrosophic model enables one to collect novel information from linguistic sources not present in a classic model | |
Using NARCBD decline the limitation on the number of new treatments that can be included in the experiment | |
The neutrosophic approach reduces the computational complexity and time, though having more complex mathematical formula | |
Neutrosophic model leads to a more powerful test than that of classic one | |
Utilizing the neutrosophic models leads to more comprehensive, precise, dependable, and enlightening outcomes |
Limitations and future research directions
This paper deals with the issue of one missing values; however, this can be extended to more than a missing value which make the problem become more complicated. For instance, in the case of two missing values, one encounter with three distinct scenarios in terms of missing cells. In the future study, the aim could be of delving this issue and extend the results to the problem of numerous missing values. Upon detecting a biased problem resulting from missing values, there are various methods to address the resolving the biasedness. Selecting a method that effectively minimizes bias is of utmost importance. The exact method proposed in this study has been limited to the ARCBD. However, in the future studies, the problem of missing values can be studied in any augmented design including augmented Latin square design, row-column design, split-plot design, etc. Finally, this study is limited to neutrosophic normality; however, in the real life, there are messy data which may not be normal. For example, data with heavy- or skewed-tail distributions are very often appeared which should be considered in the future studies.
Conclusions
The indeterminacies in data oblige ones to employ the neutrosophic type of data analysis; yet, due to the existing error and vagueness in the environment in the usual situations, applying the neutrosophic model could evermore lead to an improvement of the results. However, it must be noted that this is always an advance in methodology, making the improvement significant. Here, in this study, the proposed exact approach made the difference. Nevertheless, the progress in the computational aspect and developing a new code in the R software program should not be disregarded. This was the first attempt to consider the missing values as uncertainty matter and propose the neutrosophic model to deal with it in addition to considering the intrinsic indeterminacies in data. Moreover, all are applied for a design that is not simple, though it is very useable. Although it is conceivable that missing neutrosophic analysis using a detailed approach leads to a more efficient result, the lack of an analytical base for such an approach has prevented us from enjoying such an advantage. This study has provided a suitable environment for data analysis in such conditions. The proposed neutrosophic exact approach and obtained NANOVA, including the \( F_N \)-tests, offer greater accuracy, flexibility, applicability, and information when compared with the existing F-tests in uncertain environments.
Supplemental material
The code in R for computational materials in this manuscript can be accessed through the following link. https://github.com/AbdAita/augmented_designs.
Data availability
The data are given in the paper.
References
Allan FE, Wishart J (1930) A method of estimating the yield of a missing plot in field experimental work. J Agric Sci 20(3):399–406
Little RJ, Rubin DB (2019) Statistical analysis with missing data, vol 793. Wiley, New York
Cornish EA (1940) The estimation of missing values in incomplete randomized block experiments. Ann Eugen 10:112–18
Baird HR, Kramer CY (1960) Analysis of variance of a balanced incomplete block design with missing observations. J R Stat Soc Ser C Appl Stat 9(3):189–198
Montgomery DC (2017) Design and analysis of experiments. Wiley, New York
Coons I (1957) The analysis of covariance as a missing plot technique. Biometrics 13(3):387–405
Cochran WG (1957) Analysis of covariance: its nature and uses. Biometrics 13(3):261–281
Wilkinson GN (1958) Estimation of missing values for the analysis of incomplete data. Biometrics 14(2):257–286
Sirikasemsuk K, Leerojanaprapa K, Sirikasemsuk S (2018) Regression sum of squares of randomized complete block design with one unrecorded observation. AIP Conf Proc 2016(1):020136
Federer WT (1956) Augmented (or Hoonuiaku) designs. Biometrics Unit. Cornell Univ. Mimeo. BU-74-M
Federer WT, Crossa J (2012) I. 4 screening experimental designs for quantitative trait loci, association mapping, genotype-by environment interaction, and other investigations. Front Physiol 3:156
Federer WT (2003) Analysis for an experiment designed as augmented lattice square design. In: Handbook of formulas and software for plant geneticists and breeders, pp 283–289
Federer WT, Wolfinger RD (2003) Augmented row-column design and trend analyses. In: Handbook of formulas and software for plant geneticists and breeders, pp 291–295
Wolfinger R, Federer WT, Cordero-Brana O (1997) Recovering information in augmented designs, using SAS PROC GLM and PROC MIXED. Agron J 89(6):856–859
Kumar PS (2018) A note on ‘a new approach for solving intuitionistic fuzzy transportation problem of type-2’. Int J Logist Syst Manag 29(1):102–129
Naveed M, Riaz M, Sultan H, Ahmed N (2020) Interval valued fuzzy soft sets and algorithm of IVFSS applied to the risk analysis of prostate cancer. Int J Comput Appl 975:8887
Kumar PS (2018) PSK method for solving intuitionistic fuzzy solid transportation problems. Int J Fuzzy Syst Appl (IJFSA) 7(4):62–99
Kumar PS (2016) A simple method for solving type-2 and type-4 fuzzy transportation problems. Int J Fuzzy Logic Intell Syst 16(4):225–237
Kumar PS (2016) PSK method for solving type-1 and type-3 fuzzy transportation problems. Int J Fuzzy Syst Appl (IJFSA) 5(4):121–146
Al-shami TM (2022) (2, 1)-Fuzzy sets: properties, weighted aggregated operators and their applications to multi-criteria decision-making methods. Complex Intell Syst 9(2):1687–705
Al-shami TM, Mhemdi A (2023) Generalized frame for orthopair fuzzy sets:(m, n)-fuzzy sets and their applications to multi-criteria decision-making methods. Information 14(1):56
Al-shami TM, Alcantud JCR, Mhemdi A (2023) New generalization of fuzzy soft sets: (a, b)-fuzzy soft sets. AIMS Math 8:2995–3025
Smarandache F (2022) Neutrosophic statistics is an extension of interval statistics, while plithogenic statistics is the most general form of statistics. Infinite Study 2(2)
Smarandache F (2014) Introduction to neutrosophic statistics. Infinite Study
Aslam M (2019) A new attribute sampling plan using neutrosophic statistical interval method. Complex Intell Syst 5(4):365–370
Aslam M (2019) Neutrosophic analysis of variance: application to university students. Complex Intell Syst 5(4):403–407
AlAita A, Aslam M (2022) Analysis of covariance under neutrosophic statistics. J Stat Comput Simul 93(3):397–415
AlAita A, Talebi H, Aslam M, Al Sultan K (2023) Neutrosophic statistical analysis of split-plot designs. Soft Comput 27(12):7801–7811
Aslam M, Albassam M (2020) Presenting post hoc multiple comparison tests under neutrosophic statistics. J King Saud Univ Sci 32(6):2728–2732
Salama AA, Khaled OM, Mahfouz KM (2014) Neutrosophic correlation and simple linear regression. Neutrosophic Sets Syst 5:3–8
Nagarajan D, Broumi S, Smarandache F, Kavikumar J (2021) Analysis of neutrosophic multiple regression. Neutrosophic Sets Syst 43:44–53
Jafar MN, Saeed M, Khan KM, Alamri FS, Khalifa HAEW (2022) Distance and similarity measures using max-min operators of neutrosophic hypersoft sets with application in site selection for solid waste management systems. IEEE Access 10:11220–11235
Jafar MN, Saeed M, Saqlain M, Yang MS (2021) Trigonometric similarity measures for neutrosophic hypersoft sets with application to renewable energy source selection. IEEE Access 9:129178–129187
Jafar MN, Farooq A, Javed K, Nawaz N (2020) Similarity measures of tangent, cotangent and cosines in neutrosophic environment and their application in selection of academic programs. Infinite Study
Jafar MN, Saeed M (2021) Matrix theory for neutrosophic hypersoft set and applications in multiattributive multicriteria decision-making problems. J Math
Aslam M (2020) Design of the Bartlett and Hartley tests for homogeneity of variances under indeterminacy environment. J Taibah Univ Sci 14(1):6–10
Aslam M (2021) Chi-square test under indeterminacy: an application using pulse count data. BMC Med Res Methodol 21:1–5
Aslam M, Aldosari MS (2020) Analyzing alloy melting points data using a new Mann–Whitney test under indeterminacy. J King Saud Univ Sci 32(6):2831–2834
Sherwani RAK, Shakeel H, Awan WB, Faheem M, Aslam M (2021) Analysis of COVID-19 data using neutrosophic Kruskal Wallis H test. BMC Med Res Methodol 21(1):1–7
Sherwani RAK, Shakeel H, Saleem M, Awan WB, Aslam M, Farooq M (2021) A new neutrosophic sign test: an application to COVID-19 data. PLoS One 16(8):e0255671
Smarandache F (2021) Indeterminacy in neutrosophic theories and their applications. Infinite Study
Alhasan KFH, Smarandache F (2019). Neutrosophic Weibull distribution and neutrosophic family Weibull distribution. Infinite Study
Patro SK, Smarandache F (2016) The neutrosophic statistical distribution, more problems, more solutions. Infinite Study
Zeina MB, Miari M, Anan MT (2022) Single valued neutrosophic Kruskal–Wallis and Mann Whitney tests. Neutrosophic Sets Syst 51:948–957
AlAita A, Talebi H (2022) Neutrosophic parameters estimation and testing in augmented randomized complete block design (unpublished)
Aslam M (2022) Neutrosophic F-test for two counts of data from the poisson distribution with application in climatology. Stats 5(3):773–783
Kumar PS (2022) Computationally simple and efficient method for solving real-life mixed intuitionistic fuzzy 3D assignment problems. In J Softw Sci Comput Intell (IJSSCI) 14(1):1–42
Kumar PS (2020) Algorithms for solving the optimization problems using fuzzy and intuitionistic fuzzy set. Int J Syst Assur Eng Manag 11(1):189–222
Kumar PS (2020) Developing a new approach to solve solid assignment problems under intuitionistic fuzzy environment. Int J Fuzzy Syst Appl (IJFSA) 9(1):1–34
Kumar PS (2019) Intuitionistic fuzzy solid assignment problems: a software-based approach. Int J Syst Assur Eng Manag 10(4):661–675
Acknowledgements
The authors are deeply thankful to the editor and reviewers for their valuable suggestions to improve the quality and presentation of the paper. We are also thankful to Dr. M. M. Majidi, Professor of Plant Genetic, Breeding, and Biotechnology, College of Agriculture, Isfahan University of Technology, for delivering the original Safflower data to us.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
See Table 8.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
AlAita, A., Talebi, H. Exact neutrosophic analysis of missing value in augmented randomized complete block design. Complex Intell. Syst. 10, 509–523 (2024). https://doi.org/10.1007/s40747-023-01182-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-023-01182-5