Abstract
Blood glucose (BG) prediction is an effective approach to avoid hyper- and hypoglycemia, and achieve intelligent glucose management for patients with type 1 or serious type 2 diabetes. Recent studies have tended to adopt deep learning networks to obtain improved prediction models and more accurate prediction results, which have often required significant quantities of historical continuous glucose-monitoring (CGM) data. However, for new patients with limited historical dataset, it becomes difficult to establish an acceptable deep learning network for glucose prediction. Consequently, the goal of this study was to design a novel prediction framework with instance-based and network-based deep transfer learning for cross-subject glucose prediction based on segmented CGM time series. Taking the effects of biodiversity into consideration, dynamic time warping (DTW) was applied to determine the proper source domain dataset that shared the greatest degree of similarity for new subjects. After that, a network-based deep transfer learning method was designed with cross-domain dataset to obtain a personalized model combined with improved generalization capability. In a case study, the clinical dataset demonstrated that, with additional segmented dataset from other subjects, the proposed deep transfer learning framework achieved more accurate glucose predictions for new subjects with type 2 diabetes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus (DM) is a chronic metabolic disease, which causes a series of complications and seriously affects the patient’s health and daily life [1]. For patients with diabetes, it is necessary that they maintain their blood glucose concentration (BGC) within a target range (e.g., 70–180 mg/dL) [2]. Recently, continuous glucose-monitoring (CGM) systems [3,4,5] have been widely used in diabetes management to provide accurate and frequent dynamic glucose records in the form of time series data. By analyzing these data, deeper insight into glucose fluctuations could be obtained to provide valuable information for glucose management.
A glucose predictor would utilize the valuable CGM records to forecast the glucose level with a short-term time horizon and guide the diabetics to make proper insulin adjustments. Accurate glucose prediction is also vital for the early and proactive regulation of blood glucose before it drifts to undesirable levels. Therefore, numerous approaches, based on physical models or data-driven empirical models, have been proposed to predict glucose levels [6,7,8,9,10,11,12,13].
For physical models, it is usually difficult to provide a personalized description as the model parameters are usually impossible to estimate [14]. In contrast, data-driven empirical models provide a relatively easier way to estimate the time-varying relationships among the variables [8, 15, 16], but they have limitations in real-time prediction. The main reason is that future glucose levels are influenced by many factors such as historical trends, administered insulin, physical activity, carbohydrate intake and hormone concentration, and most of these factors are time variant and cannot be captured accurately [17, 18]. Moreover, the glucose levels of different subjects may be impacted by their different life styles and physiological reactions [19]. As a result, various prediction strategies based on complex nonlinear data-based structures only rely on CGM or a small amount of physiological status monitoring data without involving physiological variables [20,21,22].
In recent years, with the development of deep learning networks, more and more researchers have begun to apply deep learning techniques to blood glucose prediction. Mirshekarian et al. used the long short-term memory network (LSTM) with multiple physiological data to predict blood glucose, which achieved better prediction results than SVR-based methods [23]. Moreover, Li et al. proposed a convolutional recurrent neural network (CRNN) for blood glucose predictions [24]. It consisted of a multi-layer convolutional neural network (CNN) and a modified recurrent neural network (RNN) layer, in which CNN extracted the features from multi-dimensional aligned time series data, while the modified RNN layer modeled the sequence data and provided blood glucose predictions. The experimental results showed that the network performance was better than the exogenous autoregressive model (ARX), SVR and latent variable (LV) models. Aiello et al. presented a deep glucose forecasting (DGF) model that was constructed by LSTM with a full connection layer. That model had better prediction results than the average linear model and daily model predictor (DMP) [25]. Mosquera-Lopez et al. [26] developed a deep learning network that achieved superior prediction performance with the aid of huge amounts of blood glucose data (27,466 days). This model produced more accurate predictions than other machine learning approaches. The authors attributed the results to massive training data, a sufficiently complex network structure, and using different data volumes for model training. Therefore, it was reasonable to presume that deep learning networks tend to perform better when provided with sufficient training data.
Considering the significant differences in patients' physiology and behavior, many studies attempted to develop personalized deep learning models for better performance [27,28,29]. However, a common situation in clinical practice is that new patients do not have sufficient data for training a personalized model. Luo and Zhao [30] used transfer learning and incremental learning to improve the prediction accuracy of the ARX model, thus resolving the issue of insufficient data from new patients. However, that study was only concerned with traditional machine learning methods which could not benefit from the superior modeling performance of deep transfer learning.
Although various strategies for glucose prediction have been proposed in recent research studies, most of those were for patients with type 1 diabetes. Considering that type 2 diabetes accounts for a high proportion of the diabetic population and many patients with serious type 2 diabetes need to precisely manage their glucose levels, there arose a need for online glucose monitoring and prediction to provide early warning of hypoglycemia, as well as the selection of a personalized clinical medication and treatment plan [31, 32]. However, glucose dynamics may vary substantially among patients with type 2 diabetes, due to their diverse individual specificities, pathogenic factors, therapeutic schedules, and lifestyles. Therefore, there are still many challenges for the accurate prediction of glucose levels in type 2 diabetes patients and these challenges must be appropriately addressed.
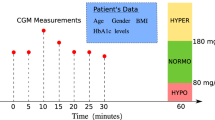
Motivated by the above considerations, a deep transfer framework to predict glucose levels of new subjects with limited historical data was proposed in this paper. By including historical segmented blood glucose data from other patients’ database, a personalized deep learning model was established to achieve dynamic short-term glucose predictions for new patients with T2D. The schematic diagram is shown in Fig. 1.
As shown in Fig. 1, the proposed deep transfer framework consisted of instances-based deep transfer and network-based deep transfer. The specific design ideas can be summarized as follows: first, a pre-trained deep learning model with superior neural network generalization ability was developed with the source domain data. Then, to guarantee the greatest level of similarity between the extended target domain and the target domain, the training dataset was augmented under the minimum dynamic time warping (DTW) distance principle [33]. Finally, the pre-trained general deep learning network was fine-tuned by data from the extended target domain to achieve the personalized prediction model. In addition to inheriting the generalization ability of the pre-trained network, the personalized model also benefited from improved prediction performance in the target domain.
The contributions of this study were as follows:
-
1.
A deep transfer learning framework was developed to establish a personalized glucose prediction model for new subjects with type 2 diabetes.
-
2.
Instance-based deep transfer learning with the minimum DTW distance was proposed to guarantee the minimal differences between the extended target domain and the original target domain.
-
3.
Based on the extended target domain, network-based deep transfer learning was developed to fine-tune the pre-trained general deep learning network to finally achieve the personalized prediction model.
-
4.
The proposed framework was applied on clinical dataset from patients with T2D, and a series of experiments were performed to verify its effectiveness and clinical acceptability.
The rest of this paper was organized as follows: Sect. 2 explains the deep transfer learning framework for glucose prediction, and introduces its related methods in detail. Section 3 describes the dataset and data pre-processing. Section 4 shows the experimental results in the clinical dataset with the proposed framework and presents necessary comparative experiments for performance analysis. Finally, Sect. 5 provides concluding comments for our study.
Deep transfer learning framework for personalized glucose prediction
Deep transfer learning is an extension of the standard transfer learning theory that addresses how to effectively transfer knowledge by deep neural network when there is insufficient data in the target domain. It can be classified into instance-based deep transfer learning, mapping-based deep transfer learning, network-based deep transfer learning, and adversarial-based deep transfer learning [34]. The unified definition of deep transfer learning is based on standard transfer learning [35], which defines a source domain \({\mathcal{D}}_{{\text{S}}}\) and a target domain \({\mathcal{D}}_{T}\) with learning tasks \({\mathcal{T}}_{{\text{S}}}\) and \({\mathcal{T}}_{{\text{T}}}\), respectively. Deep transfer learning refers to deep learning improvement of the nonlinear predictive function \(f_{{\text{T}}} ( \cdot )\) in \({\mathcal{D}}_{{\text{T}}}\) using the knowledge obtained in \({\mathcal{D}}_{{\text{S}}}\) with \({\mathcal{T}}_{{\text{S}}}\), where \({\mathcal{D}}_{{\text{S}}} \ne {\mathcal{D}}_{{\text{T}}}\), or \({\mathcal{T}}_{{\text{S}}} \ne {\mathcal{T}}_{{\text{T}}}\).
Instance-based deep transfer learning is a branch of deep transfer learning which aims to find appropriate instances from the source domain to be used as supplements for deep learning in the target domain by a well-designed weight adjustment mechanism. As an example, let us consider TrAdaBoost, which is a typical instance-based algorithm that addresses inductive transfer learning problems [36]. This algorithm assumes that data in the source and target domains have the same features and labels but different distributions. Therefore, some data in the source domain could be valuable for learning, while other data may be of no use, or may even have a destructive effect.
A small amount of data from a new subject can be regarded as the target domain \({\mathcal{D}}_{{\text{T}}}\), and glucose prediction for a new subject is equivalent to the learning task \({\mathcal{T}}_{{\text{T}}}\). The data from other subjects can be regarded as the source domain \({\mathcal{D}}_{{\text{S}}}\), where \({\mathcal{D}}_{{\text{S}}} \ne {\mathcal{D}}_{{\text{T}}}\) because of the different distributions between the subjects. Therefore, deep transfer learning can be defined as using deep learning to complete the prediction task for new subjects. The related methods of segmented data deep transfer learning for glucose prediction are described in detail in the following sections.
Instance-based transfer with minimum DTW distance
For the glucose prediction task discussed in this paper, the source domain contained the segmented glucose measurement data from multiple subjects, and the target domain only contained the segmented glucose data of a new subject. The data in the source and target domains had the same features and labels. The limited amount of data from new subjects was not enough to train a personalized deep learning prediction model, therefore, the expansion of the target domain data through instance-based transfer was the most intuitive transfer method. However, due to the differences in the physiological state and external environment between the subjects, different subjects had different distributions of glucose measurement data. Consequently, it was necessary to extract the data from the source domain that would be beneficial for transferring into the target domain. It was generally believed that it would be beneficial to learning of the target domain if the source domain data had a higher degree of similarity to the target domain. The trend and fluctuations of the sequence can be judged by the sequence shape, hence this study used dynamic time warping (DTW) distance for measurement the similarity of the glucose measurement sequences.
Dynamic time warping (DTW) is one of the most well-known pattern match techniques to determine the shape similarity between two time series [37]. Compared with other pattern match methods, DTW is a simple but effective method that has been applied in many fields [38], such as speech recognition [39, 40], computer vision and process monitoring [41, 42]. Based on a dynamic programming technique, DTW could find the minimal distance between two time series by shrinking or stretching its time dimension [33, 43].
Given two sequences \(X=({x}_{1},{x}_{2},\dots ,{x}_{m})\) and \(Y=({y}_{1}, {y}_{2},\dots ,{y}_{n})\), a distance matrix with dimension m × n was first calculated. The local distance \(d({x}_{i}, {y}_{j})\) between the corresponding elements \({x}_{i}\) and \({y}_{j}\) were measured by a distance function, such as the square Euclidean distance or Manhattan distance [44]. Then, the warping path \(W=({w}_{1},{w}_{2},\dots ,{w}_{k},\dots ,{w}_{L})\) through the distance matrix was determined by minimizing the cumulative distance, where \({w}_{k}=(i, j)\) represented the local distance \(d({x}_{i}, {y}_{j})\) on step k of warping path \(W\), and L was the length of the warping path \(W\), where \(\mathrm{ max}\left(m,n\right)\le L\le m+n-1\). The warping path \(W\) satisfied the three local constraints as follows:
-
(1)
Boundary constraints. \({w}_{1}=(\mathrm{1,1})\) and \({w}_{L}=(m,n)\).
-
(2)
Continuity. If \({w}_{k-1}=(i, j)\) and \({w}_{k}=(i{^{\prime}}, j{^{\prime}})\), then \(i-{i}^{{^{\prime}}}\le 1\) and \(j-{j}^{{^{\prime}}}\le 1\).
-
(3)
Monotonicity. If \({w}_{k-1}=(i, j)\) and \({w}_{k}=(i{^{\prime}}, j{^{\prime}})\), then \({i}^{{^{\prime}}}-i\ge 0\) and \({j}^{{^{\prime}}}-j\ge 0\).
Next, the DTW distance was designed to search the minimum warping path \({\text{DTW}}(i,j)\) by the following warping cost:
Thus, the DTW distance was a dynamic programming solution that satisfied the above three constraints.
where \(D(i,j)\) represented the DTW distance between two time series of length i and j. Finally, the DTW provided the cumulative distance to measure the similarity between the two time series. The DTW alignment is illustrated in Fig. 2, where red line shows the optimal warping path (the W in the middle). A diagonal move indicated a match between the two series, while an expansion duplicated a point in one sequence and a contraction eliminated a point.
The DTW was used to measure the level of similarity between the glucose measurement sequences in the target and source domains, and to transfer the glucose measurement sequences in the source domain that had high similarity to the target domain to construct an extended target domain. The extended target domain constructed based on the minimum DTW distance had a high degree of similarity with the original target domain, and retained the distribution of the original target domain to the greatest extent, which was desirable for representation learning of the target domain.
Network-based transfer on glucose prediction network
In network-based deep transfer learning, the pre-trained partial network in the source domain could be reused and transferred as part of the deep neural network in the target domain [34]. During the transfer process, whether the network structure needs to be modified depends on if the tasks are the same or not. For the network investigated in this study, the task was unchanged, because the glucose prediction task was performed on both the source domain and the target domain. Therefore, there was no need to modify the network structure during the transfer process, and the network structure and parameters of the pre-trained model were reused completely.
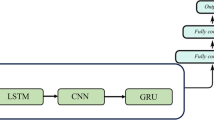
Reducing the parameters of the personalized glucose prediction model effectively shortened the update and inference time, thereby making it possible to achieve online updates and real-time predictions on mobile devices. The structure of the glucose prediction network proposed in this paper was similar to the LSTNet [45] used for time series prediction. It was mainly composed of a 1-D convolutional layer, a recurrent layer (gate recurrent unit, GRU), a batch normalization layer, and a fully connected layer, with only 427 trainable parameters as shown in Fig. 3. The 1-D convolutional layer performed preliminary feature extraction on the glucose measurement data, the recurrent layer extracted the time dependencies in the sequence, the batch normalization layer was used to improve the data distribution and training speed [46], and the fully connected layer provided the final prediction. The details of the prediction model are shown in Table 1. The inputs of this prediction model were 1-h CGM records with a sequence length of 12, and the outputs were the glucose levels a half hour or 1 h in the future.
The generalization ability of a deep learning model is largely affected by the amount and diversity of the training data. Using an extended target domain data based on the minimum DTW distance achieve a high degree of shape similarity with the target domain, but at the same time, it possibly led to the weaker generalization ability of the prediction model. The pre-trained model had a stronger generalization ability by directly training on the source domain. Subsequently, the pre-trained model was transferred to the extended target domain through network-based deep transfer learning to obtain the personalized model. The personalized model further improved the performance on the target domain while inheriting the generalization ability of the pre-trained model.
Deep transfer learning framework with segmented data
The deep transfer learning framework with segmented data is shown in Fig. 4. Initially, a deep learning model was generated for the blood glucose prediction task, and then the amount of data required for sufficient training was estimated based on the number of parameters in the model. Subsequently, DTW was used to measure the similarity between the glucose measurement sequences in the source and target domains, and enough source domain data with a high degree of similarity were transferred to the target domain to form an expanded target domain. Finally, the pre-trained model was trained on the source domain and the pre-trained model was fine-tuned on the extended target domain to obtain a personalized prediction model.
The main steps for deep transfer learning with segmented data were as follows:
-
1.
An initial deep learning model \(\theta\) was constructed for the blood glucose prediction task and experimental methods were used to estimate the minimum number of sources \(N\) required to sufficiently train the model based on the number of parameters \(n(\theta )\).
-
2.
The source domain \({\mathcal{D}}_{{\text{S}}}\)(CGM records from different subjects) was extracted from \(N - 1\) sources with the minimum DTW distance from the target domain \({\mathcal{D}}_{{\text{T}}}\) (CGM records from new subject) to construct an extended target domain \({\mathcal{D}^{\prime}}_{T}\).
-
3.
The initial model \(\theta\) was directly trained on the \(N\) sources from the source domain \({\mathcal{D}}_{{\text{S}}}\) to obtain the pre-trained model \(\theta_{{\text{S}}}\).
-
4.
The pre-trained model \(\theta_{{\text{S}}}\) was fine-tuned on the extended target domain \({\mathcal{D}^{\prime}}_{{\text{T}}}\) to obtain the personalized prediction model \(\theta_{{\text{T}}}\).
Dataset and data pre-processing
Segmented clinical dataset
This study included 908 hospitalized patients with type 2 diabetes at Department of Endocrinology and Metabolism, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital from January 2018 to the end of December 2018. Each subject wore a CGM system (Medtronic iPro2, Northridge, California) for 3–5 days for glucose measurement sampling once every 5 min. Capillary blood glucose was measured every 12 h by a SureStep blood glucose meter (LifeScan, Milpitas, CA, USA) to calibrate the CGM system. This study was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital, Shanghai, China.
To eliminate the adaptability error of subjects wearing a CGM device for the first time, only the data from the second day to the third day were extracted and each subject had 288 × 2 valid measurement points. In this study, the first 288 glucose measurements of the subjects were used as training data, and the last 288 data points were used as the testing data. The glucose measurement data from the target subject were defined as the target domain \({\mathcal{D}}_{{\text{T}}}\) and the data from other subjects served as the source domain \({\mathcal{D}}_{{\text{S}}}\). To differentiate the results and facilitate the drawing of the prediction-error grid analysis (PRED-EGA) grid, the units of the glucose measurement data were converted from mmol/L to mg/dL.
Data pre-processing
Standardizing the original data can improve the data distribution and quicken the model learning to a certain extent. The two commonly used standardization methods are min–max standardization and Z score standardization. Min–max standardization is a linear transformation of the original data and it maps the data values to [0, 1]. The Z score standardized calculation formula is shown below:
where \(x\) denoted the origin data, and \(\mu \) and \(\sigma \) were the mean and standard deviation of the original data, respectively. Z score standardization was standardized according to the mean and standard deviation of the original data. The processed data conformed to the standard normal distribution, which was beneficial to the training of the deep learning network. Therefore, Z score standardization was selected to preprocess the blood glucose data.
Results and discussion
Evaluation metrics
The efficiency and accuracy of our proposed method was demonstrated with the root mean squared error (RMSE) results in mg/dL, the mean absolute error (MAE) results in mg/dL and the mean absolute relative difference (MARD) in %, calculated by:
where \(\hat{y}_{i}\) denoted the forecasting results, \(y_{i}\) denoted the CGM measurement, and \(N\) was the total number of data points. Additionally, the prediction-error grid analysis (PRED-EGA) [47, 48] quantified the clinical acceptability of the predicted glucose levels and was used to evaluate our proposed method. The PRED-EGA was shown to be a reliable and robust method for the continuous glucose-error grid analysis [49], which evaluated the accuracy of the predictions in terms of both the values and derivatives of the predictions. The PRED-EGA included two interacting components: (1) point-EGA (P-EGA) for evaluating the accuracy of the predictions and (2) rate-EGA (R-EGA) for assessing the capability of the model to characterizes the derivative of the measurement values.
Deep transfer learning based on segmented blood glucose dataset
(1) Extended target domain
Before performing instance-base transfer, it is necessary to consider the size of the extended target domain. It is generally believed that the amount of data required to sufficiently train a model is proportional to its number of parameters, however, there is still no theoretical method to estimate the amount of data required. Therefore, experimental methods are used to estimate the data size required by the extended target domain.
The specific method employed in this study was to randomly select a certain number of subjects and use their training data as the testing dataset. One subject was randomly selected from the 908 clinical subjects in T2D to develop the predictive model with the training dataset. The performance of the model was evaluated based on the testing dataset. Next, the dataset of a subject in the training process was randomly expanded and the model was retrained and re-evaluated. This step was repeated until the model’s performance on the test dataset stabilized. In this experiment, 20 subjects were randomly selected as the test dataset, the number of subjects with maximum expansion of the training dataset was also 20, and the prediction horizon (PH) of the model was 30 min. The training settings of all the experiments in this study are shown in Table 2. The construction method of the validation dataset was the hold-out method, where the last 20% of the training data was set aside as the validation dataset. As the size of the training dataset increased, the performance of the model stabilized, as shown in Fig. 5.
The trends in Fig. 5 showed that the performance of the model on the test dataset stabilized when the number of subjects in the training dataset reached 12. However, considering the randomness of the training, a certain margin needed to be maintained, and the size of the extended target domain was finally determined to be a training dataset of 15 subjects.
(2) Joint transfer of instances and networks
The deep transfer learning framework proposed in this study was composed of instance-based transfer with DTW and network-based transfer. To evaluate the prediction performance of instance-based transfer with DTW, the models for 20 subjects in the target domain were independently trained on the extended target domain and evaluated separately. For comparison, 14 subjects were randomly selected from the source domain to construct an extended target domain with the target subjects, and the model training and evaluation were carried out in the same way. For the instance-based transfer with DTW, the network-based transfer was also used to obtain a personalized model which had improved generalization ability. Therefore, from the source domain with the target subjects excluded, 15 subjects were randomly selected to train a pre-trained model. As a result, all models of the subjects in target domain shared this pre-trained model for network-based transfer. The performance of the models generated by different transfer methods is shown in Table 3.
Table 3 shows the performance of the three transfer methods. The proposed transfer method represents that the transfer method combined with instance-based deep transfer learning with minimum DTW distance and network-based deep transfer learning. The instance transfer with DTW represented the minimum DTW distance to construct the extended target domain. The randomized instance transfer represented the random construction of the extended target domain.
The experimental results in Table 3 showed that for both PHs of 30 min and 60 min, using the minimum DTW distance for instance-based transfer improved the prediction accuracy significantly compared with using randomly selected samples. From the DTW distance calculation principle, the main factor affecting the DTW distance was the shape difference between the time series. The glucose measurement sequences that were extracted according to the minimum DTW distance with the target domain had the highest similarity versus the glucose measurement sequences that were extracted randomly. For the purposes of model learning, using glucose measurement sequences with minimum difference with the target subject made the model more sensitive to the specificity of glucose fluctuations, which would allow the model to perform better when the target subject has similar glucose fluctuations in the future.
Network-based transfer based on instance-based transfer with DTW improved the generalization ability and performance of the model. In addition, we found that the choice of the network layer for network-based transfer was crucial to the transfer results. As far as the network used in this experiment was concerned, only reusing the convolutional layer without the subsequent GRU and fully connected layer would result in negative transfer. Moreover, when the full model was fine-tuned, it was also susceptible to the influence of poorly pre-trained models and may result in negative transfer. Therefore, adopting a more reasonable network-based transfer method was very important for improving the prediction accuracy of the model. In addition to evaluating the prediction accuracy of the model, it may be more valuable to evaluate the clinical acceptability. The PRED-EGA of 20 subjects is shown in Fig. 6.
The PRED-EGA showed the clinical acceptability of the glucose estimation by the proposed method for each of the 5420 estimations from the target subjects. It was found that there was no hypoglycemia in the tested blood glucose. In the predictions for euglycemia (5183 points in total), 99.57% was accurate, 0.39% was benign, and 0.04% was error. In the predictions for hyperglycemia (237 points in total), 86.50% was accurate, 11.39% was benign, 2.11% was error. This confirmed that our proposed predictive model and transfer learning method performed well on clinical dataset. In addition, we also specifically analyzed the prediction curve of the three different deep transfer learning methods on a subject in the target domain, as shown in Fig. 7.
As shown in Fig. 7, the main source of prediction error was the rapid increase and decrease of glucose. A large prediction delay could also be observed in the model. In addition, for relatively flat glucose levels, the prediction model was prone to provide fluctuating glucose predictions, which was also the main source of prediction error. In contrast, the prediction results of the randomized instance-based transfer had too many spikes, while the prediction results of the instance-based transfer with DTW were relatively flat and the prediction performance was better. Furthermore, the use of network-based transfer improved the forecasting performance in some situations and strengthened the overall prediction results. In future work, a more appropriate deep transfer learning method may be used to improve the sensitivity and stability of the model, and further improve the performance of the deep learning model in glucose prediction.
Comparative experiment: deep transfer learning and machine learning
In addition to deep learning, segmented data can also be predicted by machine learning that requires less data. In this section, we will compare the three machine learning methods commonly used for glucose prediction, support vector regression (SVR), random forest (RF), and Gaussian process regression (GPR) with our proposed deep transfer learning method. The machine learning method only uses the training dataset of subjects in the target domain for training without extension. The machine learning model uses the encapsulated model in Python’s Sklearn library. In terms of detailed configuration, the SVR model used the radial basis function (RBF) as the kernel function, and the remaining hyperparameters were used as the default parameters. For the RF model, all the hyperparameters used default parameters. The kernel function of the GPR model was the product of ConstantKernel and RBF plus WhiteKernel. The number of restarts of the optimizer was set to 3, and the rest were default parameters. The performance of machine learning and deep transfer learning models is shown in Table 4, and the prediction accuracy of the selected subject in the target domain are shown in Fig. 8.
As can be seen in Table 4, the deep transfer learning method achieved the best prediction performance for both the 30- and 60-min PH. This result proved that the deep transfer learning method with segmented data achieved better prediction accuracy than traditional machine learning methods. It also showed that our proposed network-based transfer and instance-based transfer with minimum DTW distance is a feasible deep transfer learning method for glucose prediction. It can be seen from Fig. 8 that the prediction results of the deep transfer learning method had the best performance, and the glucose fluctuations were well captured. The prediction results of the SVR method had a high degree of fit with the raw data, were smoother than the deep transfer learning method overall, and performed better in places with minor blood glucose fluctuations. Therefore, this hybrid method using deep learning methods combined with other machine learning methods has the potential to obtain better prediction performance.
Conclusions
This study proposed a deep transfer learning framework to establish a personalized deep learning model for new patients with insufficient historical data. The minimum DTW distance was used for instance-based transfer and extended target domain, while the network-based transfer was applied to improve the comprehensive performance of the model in the target domain. The effectiveness of this method was verified through a series of blood glucose prediction comparison experiments using clinical data. One limitation of the method was that when the daily blood glucose level fluctuated significantly due to changes in the treatment plan or huge environmental changes in the clinical patients, the performance of the transfer model suffered due to changes in the target domain. To address this issue, the mean of daily differences (MODD) of blood glucose can be used, as it is a commonly used clinical indicator which reflects the consistency and stability of day-to-day blood glucose patterns [50]. Online updates can then be provided for blood glucose prediction methods with fast tracking ability for time-varying processes in this situation. Future work will also consider combining online updated machine learning methods with deep learning networks to obtain a more adaptable blood glucose prediction model.
References
American Diabetes A (2014) Diagnosis and classification of diabetes mellitus. Diabetes Care 37(Suppl 1):S81-90. https://doi.org/10.2337/dc14-S081 (Epub 2013/12/21, PubMed PMID: 24357215)
Kirchsteiger H, Jørgensen JB, Renard E, Del Re L (2015) Prediction Methods for Blood Glucose Concentration: Design, Use and Evaluation. Springer, Berlin
Klonoff DC (2005) Continuous glucose monitoring roadmap for 21st century diabetes therapy. Diabetes Care 28(5):1231–1239
Deiss D, Bolinder J, Riveline J-P, Battelino T, Bosi E, Tubiana-Rufi N et al (2006) Improved glycemic control in poorly controlled patients with type 1 diabetes using real-time continuous glucose monitoring. Diabetes Care 29(12):2730–2732
The Juvenile Diabetes Research Foundation Continuous Glucose Monitoring Study Group (2008) Continuous glucose monitoring and intensive treatment of type 1 diabetes. N Engl J Med 2008(359):1464–1476
Ozogur HN, Ozogur G, Orman Z (2020) Blood glucose level prediction for diabetes based on modified fuzzy time series and particle swarm optimization. Comput Intell-US. https://doi.org/10.1111/coin.12396 (PubMedPMID:WOS:000566826000001)
Xia Y, Rashid M, Feng J, Hobbs N, Cinar A (2020) Online glucose prediction using computationally efficient sparse kernel filtering algorithms in type-1 diabetes. IEEE Trans Control Syst Technol 28(1):3–15
Woldaregay AZ, Arsand E, Walderhaug S, Albers D, Mamykina L, Botsis T et al (2019) Data-driven modeling and prediction of blood glucose dynamics: machine learning applications in type 1 diabetes. Artif Intell Med 98:109–134. https://doi.org/10.1016/j.artmed.2019.07.007 (PubMedPMID:WOS:000488323400010)
Yu X, Turksoy K, Rashid M, Feng JY, Hobbs N, Hajizadeh I et al (2018) Model-fusion-based online glucose concentration predictions in people with type 1 diabetes. Control Eng Pract 71:129–141. https://doi.org/10.1016/j.conengprac.2017.10.013 (PubMedPMID:WOS:000424175400013)
Oviedo S, Vehi J, Calm R, Armengol J (2017) A review of personalized blood glucose prediction strategies for T1DM patients. Int J Numer Method Biomed Eng. https://doi.org/10.1002/cnm.2833 (Epub 2016/10/30, PubMed PMID: 27644067)
Georga EI, Protopappas VC, Polyzos D, Fotiadis DI (2015) Evaluation of short-term predictors of glucose concentration in type 1 diabetes combining feature ranking with regression models. Med Biol Eng Comput 53(12):1305–1318. https://doi.org/10.1007/s11517-015-1263-1 (PubMedPMID:WOS:000365753000006)
Munoz-Organero M (2020) Deep physiological model for blood glucose prediction in T1DM patients. Sensors. https://doi.org/10.3390/s20143896 (PubMed PMID: WOS:000557994000001)
Montaser E, Diez JL, Rossetti P, Rashid M, Cinar A, Bondia J (2020) Seasonal Local Models for Glucose Prediction in Type 1 Diabetes. IEEE J Biomed Health Inform 24(7):2064–2072. https://doi.org/10.1109/Jbhi.2019.2956704 (PubMedPMID:WOS:000545429400022)
Reifman J, Rajaraman S, Gribok A, Ward WK (2007) Predictive monitoring for improved management of glucose levels. J Diabetes Sci Technol 1(4):478–486
Cherkassky V, Mulier FM (2007) Learning from data: concepts, theory, and methods. Wiley, New YOrk
Araghinejad S (2013) Data-driven modeling: using MATLAB® in water resources and environmental engineering. Springer Science & Business Media, Berlin
Diabetes Research in Children Network Study Group (2005) Impact of exercise on overnight glycemic control in children with type 1 diabetes mellitus. J Pediatr 147(4):528–534
Nomura M, Fujimoto K, Higashino A, Denzumi M, Miyagawa M, Miyajima H et al (2000) Stress and coping behavior in patients with diabetes mellitus. Acta Diabetol 37(2):61–64
Brazeau A-S, Rabasa-Lhoret R, Strychar I, Mircescu H (2008) Barriers to physical activity among patients with type 1 diabetes. Diabetes Care 31(11):2108–2109
Sparacino G, Zanderigo F, Corazza S, Maran A, Facchinetti A, Cobelli C (2007) Glucose concentration can be predicted ahead in time from continuous glucose monitoring sensor time-series. IEEE Trans Biomed Eng 54(5):931–937. https://doi.org/10.1109/TBME.2006.889774 (Epub 2007/05/24, PubMed PMID: 17518291)
Reymann MP, Dorschky E, Groh BH, Martindale C, Blank P, Eskofier BM (2016) Blood glucose level prediction based on support vector regression using mobile platforms. Conf Proc IEEE Eng Med Biol Soc 2016:2990–2993. https://doi.org/10.1109/EMBC.2016.7591358 (Epub 2017/03/09, PubMed PMID: 28268941)
Pérez-Gandía C, Facchinetti A, Sparacino G, Cobelli C, Gómez E, Rigla M et al (2010) Artificial neural network algorithm for online glucose prediction from continuous glucose monitoring. Diabetes Technol Ther 12(1):81–88
Mirshekarian S, Bunescu R, Marling C, Schwartz F (2017) Using LSTMs to learn physiological models of blood glucose behavior. Conf Proc IEEE Eng Med Biol Soc 2017:2887–2891. https://doi.org/10.1109/EMBC.2017.8037460 (Epub 2017/10/25, PubMed PMID: 29060501)
Li K, Daniels J, Liu C, Herrero P, Georgiou P (2020) Convolutional recurrent neural networks for glucose prediction. IEEE J Biomed Health Inform 24(2):603–613. https://doi.org/10.1109/JBHI.2019.2908488 (Epub 2019/04/05, PubMed PMID: 30946685)
Aiello EM, Lisanti G, Magni L, Musci M, Toffanin C (2020) Therapy-driven deep glucose forecasting. Eng Appl Artif Intel. https://doi.org/10.1016/j.engappai.2019.103255
Mosquera-Lopez C, Dodier R, Tyler N, Resalat N, Jacobs P (2019) Leveraging a big dataset to develop a recurrent neural network to predict adverse glycemic events in type 1 diabetes. IEEE J Biomed Health Inform. https://doi.org/10.1109/JBHI.2019.2911701 (Epub 2019/04/19, PubMed PMID: 30998484)
Eberle C, Ament C (2012) Real-time state estimation and long-term model adaptation: a two-sided approach toward personalized diagnosis of glucose and insulin levels. J Diabetes Sci Technol 6(5):1148–1158. https://doi.org/10.1177/193229681200600520 (Epub 2012/10/16, PubMed PMID: 23063042; PubMed Central PMCID: PMCPMC3570850)
Balakrishnan NP, Rangaiah GP, Samavedham L (2012) Personalized blood glucose models for exercise, meal and insulin interventions in type 1 diabetic children. Conf Proc IEEE Eng Med Biol Soc 2012:1250–1253. https://doi.org/10.1109/EMBC.2012.6346164 (Epub 2013/02/01, PubMed PMID: 23366125)
Capel I, Rigla M, Garcia-Saez G, Rodriguez-Herrero A, Pons B, Subias D et al (2014) Artificial pancreas using a personalized rule-based controller achieves overnight normoglycemia in patients with type 1 diabetes. Diabetes Technol Ther 16(3):172–179. https://doi.org/10.1089/dia.2013.0229 (Epub 2013/10/25, PubMed PMID: 24152323; PubMed Central PMCID: PMCPMC3934437)
Luo S, Zhao C (2019) Transfer and incremental learning method for blood glucose prediction of new subjects with type 1 diabetes. In: 2019 12th Asian control conference (ASCC); Kitakyushu-shi, Japan, pp 73–78
Chemlal S, Colberg S, Satin-Smith M, Gyuricsko E, Hubbard T, Scerbo MW et al (2011) Blood glucose individualized prediction for type 2 diabetes using iPhone application. Northeast Bioengin C (PubMed PMID: WOS:000297284900197)
Franc S, Dardari D, Riveline JP, Charpentier G (2008) Prediction of postprandial blood glucose level according to the amount of carbohydrates consumed during a meal in type 2 diabetes patients. Diabetes 57:A552–A553 (PubMed PMID: WOS:000256612002502)
Narita H, Sawamura Y, Hayashi A (2009) DTW-distance based kernel for time series data. IEICE Trans Inf Syst E92d(1):51–58. https://doi.org/10.1587/transinf.E92.D.51 (PubMed PMID: WOS:000263079200007)
Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C (2018) A survey on deep transfer learning. arXiv preprint. arXiv: 1808.01974v1. https://doi.org/10.1007/978-3-030-01424-7_27.
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359. https://doi.org/10.1109/tkde.2009.191
Dai W, Yang Q, Xue G-R, Yu Y (2007) Boosting for transfer learning. In: Proceedings of the 24th international conference on machine learning. Association for Computing Machinery, Corvalis, Oregon, USA, pp 193–200
Deng HQ, Chen WF, Shen Q, Ma AJ, Yuen PC, Feng GC (2020) Invariant subspace learning for time series data based on dynamic time warping distance. Pattern Recogn. https://doi.org/10.1016/j.patcog.2020.107210 (PubMed PMID: WOS:000525825100019)
Kate RJ (2016) Using dynamic time warping distances as features for improved time series classification. Data Min Knowl Disc 30(2):283–312. https://doi.org/10.1007/s10618-015-0418-x (PubMedPMID:WOS:000370157600001)
Chiu CC, Shanblatt MA (1995) Human-like dynamic-programming neural networks for dynamic time warping speech recognition. Int J Neural Syst 6(1):79–89. https://doi.org/10.1142/S012906579500007x (PubMedPMID:WOS:A1995RB89000007)
Axelrod S, Maison B (2004) Combination of hidden Markov models with dynamic time warping for speech recognition. In: 2004 IEEE international conference on acoustics, speech, and signal processing, vol I, Proceedings, pp 173–6 (PubMed PMID: WOS:000222173500044)
Zhang YH, Zhao H (2020) Land-use and land-cover change detection using dynamic time warping-based time series clustering method. Can J Remote Sens 46(1):67–83. https://doi.org/10.1080/07038992.2020.1740083 (PubMedPMID:WOS:000524196500001)
Tian Y, Wang ZL, Lu C (2019) Self-adaptive bearing fault diagnosis based on permutation entropy and manifold-based dynamic time warping. Mech Syst Signal Process 114:658–673. https://doi.org/10.1016/j.ymssp.2016.04.028 (PubMedPMID:WOS:000447112700038)
Salvador S, Chan P (2007) Toward accurate dynamic time warping in linear time and space. Intell Data Anal 11(5):561–580. https://doi.org/10.3233/ida-2007-11508
Yu Z, Niu Z, Tang W, Wu Q (2019) Deep learning for daily peak load forecasting–a novel gated recurrent neural network combining dynamic time warping. IEEE Access 7:17184–17194. https://doi.org/10.1109/access.2019.2895604
Lai G, Chang W-C, Yang Y, Liu H (2018) Modeling long- and short-term temporal patterns with deep neural networks. In: The 41st international ACM SIGIR conference on research and development in information retrieval, pp 95–104
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint. ar xiv:1502.03167
Sivananthan S, Naumova V, Man CD, Facchinetti A, Renard E, Cobelli C et al (2011) Assessment of blood glucose predictors: the prediction-error grid analysis. Diabetes Technol Ther 13(8):787–796. https://doi.org/10.1089/dia.2011.0033 (Epub 2011/05/27, PubMed PMID: 21612393)
Kovatchev BP, Gonder-Frederick LA, Cox DJ, Clarke WL (2004) Evaluating the accuracy of continuous glucose-monitoring sensors: continuous glucose-error grid analysis illustrated by TheraSense Freestyle Navigator data. Diabetes Care 27(8):1922–1928. https://doi.org/10.2337/diacare.27.8.1922 (Epub 2004/07/28, PubMed PMID: 15277418)
Clarke WL, Cox D, Gonder-Frederick LA, Carter W, Pohl SL (1987) Evaluating clinical accuracy of systems for self-monitoring of blood glucose. Diabetes Care 10(5):622–628. https://doi.org/10.2337/diacare.10.5.622 (Epub 1987/09/01, PubMed PMID: 3677983)
Alemzadeh R, Loppnow C, Kirby M, Parton E, Haas P (2003) Glucose sensor evaluation of glycemic instability in pediatric type 1 diabetes mellitus. Diabetes Technol Ther 5(2):167–173
Acknowledgements
This work was funded by the National Natural Science Foundation of China (61903071, 61973067), the National Key R&D Program of China (2018YFC2001004) and the Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (20161430).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No competing financial interests exist.
Ethics approval
The study was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (2020-KY-024).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, X., Yang, T., Lu, J. et al. Deep transfer learning: a novel glucose prediction framework for new subjects with type 2 diabetes. Complex Intell. Syst. 8, 1875–1887 (2022). https://doi.org/10.1007/s40747-021-00360-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00360-7