Abstract
Brain tumor is a group of anomalous cells. The brain is enclosed in a more rigid skull. The abnormal cell grows and initiates a tumor. Detection of tumor is a complicated task due to irregular tumor shape. The proposed technique contains four phases, which are lesion enhancement, feature extraction and selection for classification, localization, and segmentation. The magnetic resonance imaging (MRI) images are noisy due to certain factors, such as image acquisition, and fluctuation in magnetic field coil. Therefore, a homomorphic wavelet filer is used for noise reduction. Later, extracted features from inceptionv3 pre-trained model and informative features are selected using a non-dominated sorted genetic algorithm (NSGA). The optimized features are forwarded for classification after which tumor slices are passed to YOLOv2-inceptionv3 model designed for the localization of tumor region such that features are extracted from depth-concatenation (mixed-4) layer of inceptionv3 model and supplied to YOLOv2. The localized images are passed to McCulloch's Kapur entropy method to segment actual tumor region. Finally, the proposed technique is validated on three benchmark databases BRATS 2018, BRATS 2019, and BRATS 2020 for tumor detection. The proposed method achieved greater than 0.90 prediction scores in localization, segmentation and classification of brain lesions. Moreover, classification and segmentation outcomes are superior as compared to existing methods.
Similar content being viewed by others
Introduction
The tumor is a mass of irregular cells called the primary brain tumor inside the brain. The common symptoms of brain tumors are headaches, seizures, difficulties in speech, vomiting, imbalance problem, sensation loss, changes in behavior, and personality [58]. In America, 700,000 persons are suffering from brain tumor, and expected to increase to more than 79,000 by the end of 2020. Among these, 25,000 may suffer from malignant and remaining from non-malignant tumor [15]. Glioma is a predominant form of brain tumor, broken into low- and high-grade brain tumors. such that high grade is more aggressive as compared to low grade [13]. MRI is utilized to examine anatomical body structure [20, 32], which is widely used for the detection of brain tumors. An error-prone and more exhaustive activity is manual diagnosis of brain tumors using MRI. Therefore, automated approaches are used for anomalous detection which is helpful for accurate and fast detection [2, 8,9,10, 42,43,44,45]. Nowadays, several researchers are focused on different imaging sequences of MRI to analyze the tumor region [9, 61, 66]. Several techniques are introduced in literature based on clustering [19, 31, 47] and super pixels [54] for brain tumor detection. Appropriate extraction of features and optimization is a difficult task i.e., [56], particle swarm optimization (PSO) [31], local binary patterns (LBP), and histogram features [1, 59] are utilized for the classification of tumor. The existing approaches have failed for the detection of more than one small volume of tumor per MRI slices [29]. These methods detect tumors on only Flair imaging modality such that SVM has been utilized for classification that performed better on small data. Hence, there is still a need of improved techniques for tumors detection on different views, such as saggital, coronal, and axial from large-scale imaging data [5, 14]. Keeping this in view, an improved approach is presented in this article for classification, localization, and segmentation of glioma lesions. The major article contribution is opted as follows:
-
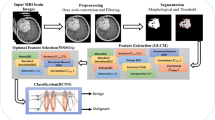
The homomorphic wavelet filer is applied on input MRI images for noise removal and passed to the pre-trained inceptionv3 model for feature extraction, where optimum features are selected using NSGA.
-
After classification, infected region is localized based on YOLOv2-inceptionv3 model, where deep features are extracted using depth-concatenation (mixed-5) layer and passed to YOLOv2 model.
-
McCulloch's Kapur entropy is applied to localized images for 3D-segmentation of tumor region. The segmentation outcome is also validated with truth annotated images to confirm the method's effectiveness.
The remaining manuscript is divided in different sections i.e., related work is in “Related work”, and proposed work with respected results are presented in “Proposed methodology” and “Results and discussion”, respectively.
Related work
Extensive work has been done for brain tumor detection [11]. Enhancement is a more vital task for noise reduction that aids in the improvement of segmentation. Wavelet filter [50], median filter [7], Gaussian filter [52], PDDF filter, FNLM filter [49], and high-pass filter [7] are used in pre-processing step. Pereira et al. [41] applied CNN with 3 kernel sizes and obtained 0.88, 0.83, 0.77 dice scores of complete, enhance, and non-enhanced tumor regions, respectively. Sauwen et al. [48] proposed different methodologies to analyze tumor segmentation results [26]. Goswami and Bhaiya [6] presented a hybrid framework consisting of fuzzy logic and neural network for tumor detection and classification [51]. A semi-automatic method with spatial features is applied for tumor detection [24]. Different clustering approaches (K-means [8], PSO, MFKM) are used for the segmentation of tumor [60]. Watershed is utilized with GLCM for features extraction and supplied to SVM [53] for multi-fractals classification with a higher precision rate. The transfer learning models are widely utilized to classify the tumor region, such as Alex-net, Google Net, and VGG-16. Two different types of neural networks are trained on augmented input images for brain lesions classification [52]. The pre-trained AlexNet has been utilized for glioma detection for the prediction of patient’s survival rate [53]. CNN model is trained on brain imaging data and classified input data into five classes, such as multiform glioma, astrocytoma, shapeless tumor, normal brain tissues, and oligodendroglioma [6]. M-net segmentation model has been utilized for features extraction and fed into the pre-trained VGG-16 for the classification of three different types of the tumor [63]. Fuzzy-c-means has been applied for segmentation followed by DWT features extraction and suitable features selection by PCA for classification [35]. Capsule Networks (CapsNets) has been utilized [3]. 3-D CNN architecture has been utilized for glioma classification into different grades, such as low and high [23]. 2-D-CNN has been used for increasing the precision rate of glioma classification [21, 22]. Deep CNN network has been applied for glioma classification. 3D-Unetwork has been used for glioma detection in which average global pooling layer is used for features mapping followed through 1 × 1 cascade convolutional work as FC layer [7]. A CNN model is utilized for deep features extraction and informative features selection using GA for glioma classification [12]. While comprehensive tumor detection and classification work has been performed, but still accurate tumor detection is a challenging task and has room for improvement. Therefore, this research work provides an improved approach for classification, localization, and segmentation of brain tumor.
Proposed methodology
The proposed method has four primary steps: (1) enhancement, (2) classification, (3) localization, and 4) segmentation as illustrated in Fig. 1 such that input images are enhanced using homomorphic wavelet filer and classified using extracted deep features from inceptionv3. The classified images are localized through the proposed YOLOv2-inceptionv3 and segmented based on Kapur entropy.
Noise elimination using homomorphic wavelet filter
The images acquired from MRI protocol having adversative situations might be contaminated due to noise that degrades the disease detection rate. Several filters are presented for noise removal. These filters depend on noise type included in the images. Wavelet transform is used to represent the images into frequency domain. In this process, image decomposition is performed to process the image into high–high (HH), low–high (LH), and high–low (HL) bands. This research investigates a homomorphic wavelet filter decomposition to eliminate speckle noise that is mathematically expressed as follows:
The noise removal process using a homomorphic filter with wavelet decomposition is visualized in Fig. 2 such that image is decomposed into 04 bands HL, LH, HH, and LH-HH. The HH band improves the image quality as compared to other bands like HL, LH, and LH-HH. Thus, for further processing, HH band is utilized to perform accurate segmentation.
Extracted deep features using pre-trained inceptionv3 architecture
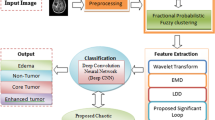
Deep learning is widely utilized in artificial intelligence applications, such as speech recognition and computer vision. However, with more interest in the area of deep learning, classification into corresponding categories is a major problem. This problem might be solved through transfer learning because accurate models and architecture are built in in a time-saving manner. In this process, learning is performed through already learned patterns to solve different problems instead of using features learning from scratch. Transfer learning uses pre-trained models that are learned on huge amount of data for problem-solving. Thus, this work utilizes an inceptionv3 pre-trained transfer learning model [55] for features learning which consists of 01 image, 094 Convolutional (Conv), 094 batch-normalization (bn), 094 ReLU, 14 max-pooling, 015 depth concatenation, fully connected layers, and softmax with cross-entropy function. The features are extracted from fully connected layers named as prediction and further passed to NSGA [18] for improved features selection as displayed in Fig. 3.
Features Selection
A deep feature vector (1 × 1000) is obtained using inceptionv3 network. The features of engineering are performed to select optimum feature vector by applying NSGA II. The parameters of NSGA as are discussed in Table 1.
Localization using YOLOv2-inceptionv3 model
YOLOv2-inceptionv3 model with 174 layers is proposed to localize tumor region such that there is 165 layers of inceptionv3 with 01 input, 50 Conv, 50 bn, 50 activations ReLU, 06 mixed (depth concatenation) 03 max-pooling, 05 average pooling, and 09 layers of tinyYOLOv2 [46] model. The optimum hyper-parameters are discussed in Table 2.
The proposed model more accurately localizes tumor region as illustrated in Fig. 4.
YOLOv2 model optimized MSE loss among predicted bounding and ground truth boxes. The model training is performed on three different types of losses, such as localization, confidence, and classification. Among the expected and ground truth boxes, localization loss computes error using location, estimated box size, and ground truth. The confidence loss is utilized to compute objectiveness error with detected object in jth bounded box of grid i cell. The classification loss is used to calculate probability across each class of grid cell i. The mathematical formulation of these parameters is expressed as:
Here, s represents grid cell, p denotes probability, w1, w2,w3 and w4 show weights, gc presents grid cell, \(\left( {\widehat{{x_{i} }},\widehat{{y_{i} }}} \right)\) denotes center of bounding box, \((x_{i} ,y_{i} ) \) shows center of ground truth. \(({\text{width}}_{{\text{i}}} , {\text{height}}_{{\text{i}}} )\) signifies width and height of bounding box and \((\widehat{{{\text{width}}}}_{ii} ,\,\widehat{{{\text{height}}}})\) presents width and height of ground truth.
Lesion segmentation
A key challenge in medical images is variability in medical data. In human anatomy, variations occur in different modalities including X-ray, MRI, CT, and PET, etc. The segmentation region is used to analyze the disease severity levels. In the proposed method, McCulloch's Kapur entropy method [28] is utilized for tumor segmentation. In this method, probability of intensity values distribution is measured from the foreground and background regions after which entropy is calculated separately from both regions. The optimum value of threshold is applied to increase the sum of their entropies. The Kapur entropy is mathematically expressed as:
Here
Figure 5 visualizes the effects of tumor segmentation.
Results and discussion
The method is evaluated on BRATS series including 2018, 2019, and 2020 [16, 30, 33]. BRATS 2018 contains 266 MRI patients with 191 high and 75 low glioma grade, BRATS 2019 composes of 285 patients, and BRATS 2020 has 335 patients such that each patient contains 155 slices. The detailed description of benchmark databases is illustrated in Fig. 6.
The 0.5 hold-out validation approach is utilized for tumor slices classification, where half data are used for training and remaining for validation. The summary of classified images is given in Table 3.
The proposed work is evaluated on experiments implemented on MATLAB 2020-Ra toolbox with 2070 Nvidia Graphic Card and Gamming Laptop G5-5500 to validate the enhancement method, classification approach, localization technique, and segmentation method, respectively.
Experiment#1
In this experiment, the enhancement technique is evaluated in terms of different performance metrics, such as SSIM, MSE, and PSNR. The enhancements results are mentioned in Table 4 as well as visually presented in Fig. 7.
In Fig. 7, quantitative results are computed in terms of MSE, SNR, and PSNR using four bands, such as HL, LH, HH, and LH-HH. In this procedure, 80.46 PSNR, 70.70 SNR, 4.8 MSE on LH band, 83.43 PSNR, 73.68 SNR, 2.9 MSE on LH band, 87.68 PSNR, 77.92 SNR, 1.1 MSE on HH band, and 84.46 PSNR, 74.71 SNR, 2.3 MSE are achieved on LH-HH band, hence concluding that HH band showed highest measures. Ten sample images are taken to compute the metrics as shown in Table 4.
The results in Table 4 depict that proposed method attained maximum 89.4096815346192 PSNR, 78.0342291390692 SNR, and 0.000369981932768593 MSE. The overall performance is represented in Fig. 8.
Experiment#2
In experiment#2, tumor predictions are done on 0.5 hold-out validation that is mentioned in Tables 5, 6, 7, 8. The method classified brain images (normal (0) and abnormal (1)) as shown in confusion matrices in Fig. 9. Figure 10 shows ROC on BRATS datasets with maximum 1.00 AUC and minimum 0.98 AUC.
In terms of performance metrics, BRATS 2018 obtained 0.0000 FNR while BRATS 2020 achieved 0.0138 FNR.
Table 6 shows analysis of applying different classifiers to final features vector, where DT achieves 95% ACC, 97% SP, 88% SE, 85% PPV, 0.0300 FPR, and 0.1132 FNR. On discriminant analysis, quadratic kernel obtains highest results in comparison with linear kernel, such as 98% ACC on quadratic and 97% on linear kernel of LDA. On SVM, quadratic kernel attains 93% PPV and linear kernel shows 92% PPV.
The results in Table 7 show that DT achieves 94% ACC, while discriminant analysis shows 98% ACC using quadratic and 97% ACC using linear kernel. In geometrical family, SVM achieves 98% ACC on linear and 99% ACC on quadratic and cubic kernels.
From the results in Tables 5, 6, 7, 8, SVM (cubic kernel) achieves maximum 0.9891 ACC whereas minimum 0.9563 ACC is obtained using DT on BRATS 2018. Likewise, on BRATS 2019, SVM (cubic kernel) attains maximum 0.9970 ACC and minimum 0.9421 ACC is obtained using DT. On BRATS 2020, SVM (quadratic kernel) shows maximum 0.9939 ACC while minimum 0.9329 ACC is attained with DT. Finally, it is observed that SVM performs better than other classifiers. Proposed method results comparison is stated in Table 9.
Table 9 shows the results comparison with existing work, such as [4, 37, 65, 67], such that 94% SE and 95% SE are obtained on BRATS 2018 while 96% SE is attained on BRATS 2019 datasets, respectively. However, SE of 100% and 99% are shown on BRATS 2018 and BRATS 2019 datasets, respectively, using proposed method.
Experiment#3
In this experiment, YOLOv2-inceptionv3 model is validated on performance metrics, such as mAP and IoU, as shown in Table 10 such that proposed method achieved mAP of 0.98, 0.99 and 1.00 on BRATS 2018, 2019 and 2020, respectively. The recommended approach localizes tumor region with highest confidence scores presented in the Fig. 11.
Experiment#4
In this experiment, localized images are segmented to analyze actual infected region more precisiely. The mathmatical formulation of segmentation measures, such as dice and jaccard index, is defined as:
In this experiment, localized images are segmented to analyze the actual infected region more precisiely. The numerical computed results are also discussed in Table 11.
From the results in Table 11, it is observed that on HGG glioma, maximum 1.00 (dice, Jaccard index) and minimum 0.99, 0.98 (dice, Jaccard index) are obtained. On LGG, maximum 1.00 and minimum 0.99 (dice, Jaccard index) are achieved, respectively. The average segmentation outcomes on BRATS series are listed in Table 12.
Table 12 shows that proposed framework achieved dice of 0.98, 0.96 and 0.97 on BRATS 2018, 2019 and 2020 datasets. The segmentation results on HGG and LGG are visualized in Figs. 12, 13, 14.
The results comparison is given in Table 13.
The proposed segmented results are compared with eight recent published works, such as [27, 34, 36, 38, 40, 57, 62]. The existing methods achieved maximum 0.82 dice score on 2018 BRATS, 0.89 dice score on 2019 BRATS and 0.84 dice score on BRATS 2020 datasets. In comparison with existing methods, presented framework achieved 0.98, 0.96 and 0.97 scores on BRATS 2018, 2019 and 2020 databases, respectively.
Conclusion
The comprehensive experiments are conducted to evaluate the proposed method performance using recent TOP MICCAI Challenging datasets. The enhancement results are improved using homomorphic wavelet decomposition analysis and achieved 89.4 PSNR, 78.03 SNR, and 0.00036 MSE. The pixel-wise (segmentation results) depict 1.00 DSC. The softmax as well as multiple classifiers (KNN, SVM, LDA, ensemble and DT) with 0.5 hold-out is used to classify healthy and unhealthy slices. Finally, it is concluded that softmax provided competitive outcomes with 0.99 ACC as compared to other classifiers. These evaluation results prove that this research provided help to classify tumor accurately. After classification, the classified tumor images are localized using proposed YOLOv2-inceptionv3 model. The proposed model more accurately detected the tumor region in terms of mAP 0.98, 0.99 and 1.00 on BRATS 2018, 2019 and 2020 databases, respectively. The localized region is segmented using proposed Kapur entropy method. The experimental results conclude that proposed approach achieved competitive results than the recent published work. The improved hybrid approach can be utilized in real-time applications to diagnose brain tumor at a premature stage. This research will be further expanded in future for the study of brain tumors using algorithms of quantum computation.
References
Abbasi S, Tajeripour F (2017) Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient. Neurocomputing 219:526–535
Acharya UR, Fernandes SL, WeiKoh JE, Ciaccio EJ, Fabell MKM, Tanik UJ, Rajinikanth V, Yeong CH (2019) Automated detection of Alzheimer’s disease using brain MRI images–a study with various feature extraction techniques. J Med Syst 43:302
Afshar P, Mohammadi A, Plataniotis KN (2018) Brain tumor type classification via capsule networks. In: 2018 25th IEEE International Conference on Image Processing (ICIP), IEEE, pp 3129–3133
Ahuja S, Panigrahi B, Gandhi T (2020) Transfer Learning Based Brain Tumor Detection and Segmentation using Superpixel Technique. In: 2020 International Conference on Contemporary Computing and Applications (IC3A), IEEE, pp 244–249
Al-Saffar ZA, Yildirim T (2021) A hybrid approach based on Multiple Eigenvalues Selection (MES) for the automated grading of a brain tumor using MRI. Comput Meth Progr Biomed 201:105945
Alhassan AM, Zainon WMNW (2020) BAT algorithm with fuzzy C-ordered means (BAFCOM) clustering segmentation and enhanced capsule networks (ECN) for brain cancer MRI images classification. IEEE Access 8:201741–201751
Amin J, Sharif M, Gul N, Raza M, Anjum MA, Nisar MW, Bukhari SACJJoMS, (2020) Brain tumor detection by using stacked autoencoders in deep learning. J Med Syst 44:32
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit Lett 139:118–127
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: Deep convolutional neural networks. Futur Gener Comput Syst 87:290–297
Amin J, Sharif M, Yasmin M, Saba T, Anjum MA, Fernandes SL (2019) A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning. J Med Syst 43:326
Amin J, Sharif M, Yasmin M, Saba T, Raza MJMT (2020) Applications Use of machine intelligence to conduct analysis of human brain data for detection of abnormalities in its cognitive functions. Multimed Tools Appl 79:10955–10973
Anaraki AK, Ayati M, Kazemi F (2019) Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern Biomed Eng 39:63–74
Angelini ED, Clatz O, Mandonnet E, Konukoglu E, Capelle L, Duffau H (2007) Glioma dynamics and computational models: a review of segmentation, registration, and in silico growth algorithms and their clinical applications. Curr Med Imaging Rev 3:262–276
Ayadi W, Elhamzi W, Charfi I, Atri M (2021) Deep CNN for brain tumor classification. Neural Process Lett. https://doi.org/10.1007/s11063-020-10398-2
Bahadure NB, Ray AK, Thethi HP (2018) Comparative approach of MRI-based brain tumor segmentation and classification using genetic algorithm. J Digit Imaging. https://doi.org/10.1007/s10278-018-0050-6
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C (2017) Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 4:170117
Breiman L, Friedman JH, Olshen RA, Stone CJ (1984) Classification and regression trees. Wadsworth, Belmont, CA
Conn AR, Gould NI, Toint PL (1991) A globally convergent augmented Lagrangian algorithm for optimization with general constraints and simple bounds. SIAM J Numeric Anal 28:545–572
David DS, Jayachandran A (2018) Robust classification of brain tumor in MRI images using salient structure descriptor and RBF kernel-SVM. TAGA J Graphic Technol 14(64):718–737
DeAngelis LM (2001) Brain tumors. N Engl J Med 344:114–123
Decuyper M, Bonte S, Van Holen R (2018) Binary glioma grading: radiomics versus pre-trained CNN features. International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 498–505
Ge C, Gu IY-H, Jakola AS, Yang J (2018) Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE, pp 5894–5897
Ge C, Qu Q, Gu IY-H, Jakola AS (2018) 3D multi-scale convolutional networks for glioma grading using MR images. In: 2018 25th IEEE International Conference on Image Processing (ICIP) IEEE, pp 141–145
Gordillo N, Montseny E, Sobrevilla PJ (2013) State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging 31:1426–1438
Hastie T, Tibshirani R, Friedman J (2009) The elements of statistical learning: data mining, inference, and prediction. Springer Science and Business Media, Berlin
Havaei M, Larochelle H, Poulin P, Jodoin P-M (2016) Within-brain classification for brain tumor segmentation. Int J Comput Assist Radiol Surg 11:777–788
Henry T, Carre A, Lerousseau M, Estienne T, Robert C, Paragios N, Deutsch E (2020) Top 10 BraTS 2020 challenge solution: brain tumor segmentation with self-ensembled, deeply-supervised 3D-Unet like neural networks. arXiv:2011.01045
Kapur JN, Sahoo PK, Wong AK (1985) A new method for gray-level picture thresholding using the entropy of the histogram. Comput Vis Graph Image Process 29:273–285
Khalil HA, Darwish S, Ibrahim YM, Hassan OF (2020) 3D-MRI brain tumor detection model using modified version of level set segmentation based on dragonfly algorithm. Symmetry 12:1256
Kistler M, Bonaretti S, Pfahrer M, Niklaus R, Büchler P (2013) The virtual skeleton database: an open access repository for biomedical research and collaboration. J Med Internet Res 15:e245
Lahmiri S (2017) Glioma detection based on multi-fractal features of segmented brain MRI by particle swarm optimization techniques. Biomed Signal Process Control 31:148–155
Liang Z-P, Lauterbur PC (2000) Principles of magnetic resonance imaging: a signal processing perspective. SPIE Optical Engineering Press, London
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024
Messaoudi H, Belaid A, Allaoui ML, Zetout A, Allili MS, Tliba S, Salem DB, Conze PH (2020) Efficient embedding network for 3D brain tumor segmentation. arXiv:2011.11052
Mohsen H, El-Dahshan E-SA, El-Horbaty E-SM, Salem A-BM (2018) Classification using deep learning neural networks for brain tumors. Future Comput Inform J 3:68–71
Murugesan GK, Nalawade S, Ganesh C, Wagner B, Fang FY, Fei B, Madhuranthakam AJ, Maldjian JA (2019) Multidimensional and multiresolution ensemble networks for brain tumor segmentation. International MICCAI brainlesion workshop. Springer, Berlin, pp 148–157
Narmatha C, Eljack SM, Tuka AARM, Manimurugan S, Mustafa M (2020) A hybrid fuzzy brain-storm optimization algorithm for the classification of brain tumor MRI images. J Ambient Intellig Humaniz Comput. https://doi.org/10.1007/s12652-020-02470-5
Nguyen HT, Le TT, Nguyen TV, Nguyen NT (2020) Enhancing MRI brain tumor segmentation with an additional classification network.
Pei L, Vidyaratne L, Hsu W-W, Rahman MM, Iftekharuddin KM (2019) Brain tumor classification using 3d convolutional neural network. International MICCAI brainlesion workshop. Springer, Berlin, pp 335–342
Pei L, Vidyaratne L, Rahman MM, Iftekharuddin KM (2020) Context aware deep learning for brain tumor segmentation, subtype classification, and survival prediction using radiology images. Sci Rep 10:1–11
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35:1240–1251
Raja NSM, Fernandes S, Dey N, Satapathy SC, Rajinikanth V (2018) Contrast enhanced medical MRI evaluation using Tsallis entropy and region growing segmentation. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-018-0854-8
Rajinikanth V, Fernandes SL, Bhushan B, Sunder NR (2018) Segmentation and analysis of brain tumor using Tsallis entropy and regularised level set. Proceedings of 2nd international conference on micro-electronics, electromagnetics and telecommunications. Springer, Berlin, pp 313–321
Rajinikanth V, Satapathy SC, Fernandes SL, Nachiappan S (2017) Entropy based segmentation of tumor from brain MR images–a study with teaching learning based optimization. Pattern Recogn Lett 94:87–95
Rajinikanth V, Thanaraj KP, Satapathy SC, Fernandes SL, Dey N (2019) Shannon’s entropy and watershed algorithm based technique to inspect ischemic stroke wound. Smart intelligent computing and applications. Springer, Berlin, pp 23–31
Redmon J, Farhadi A (2017) YOLO9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7263–7271
Samanta AK, Khan AA (2018) Computer aided diagnostic system for automatic detection of brain tumor through MRI using clustering based segmentation technique and SVM classifier. International conference on advanced machine learning technologies and applications. Springer, Berlin, pp 343–351
Sauwen N, Acou M, Van Cauter S, Sima D, Veraart J, Maes F, Himmelreich U, Achten E, Van Huffel S (2016) Comparison of unsupervised classification methods for brain tumor segmentation using multi-parametric MRI. Neuroimage 12:753–764
Sharif M, Amin J, Nisar MW, Anjum MA, Muhammad N, Shad SAJCSR (2020) A unified patch based method for brain tumor detection using features fusion. Cognit Syst Res 59:273–286
Sharma M, Patel S, Acharya URJPRL (2020) Automated detection of abnormal EEG signals using localized wavelet filter banks. Pattern Recognit Lett. https://doi.org/10.1016/j.patrec.2020.03.009
Sheela CJJ, Suganthi GJMT (2020) Morphological edge detection and brain tumor segmentation in Magnetic Resonance (MR) images based on region growing and performance evaluation of modified Fuzzy C-Means (FCM) algorithm. Multimed Tools Appl. https://doi.org/10.1007/s11042-020-08636-9
Shukla M, Sharma KK (2020) A comparative study to detect tumor in brain MRI images using clustering algorithms. In: 2020 2nd International Conference on Innovative Mechanisms for Industry Applications (ICIMIA), IEEE, pp 773–777
Singh NP, Dixit S, Akshaya A, Khodanpur B (2017) Gradient magnitude based watershed segmentation for brain tumor segmentation and classification. Proceedings of the 5th international conference on frontiers in intelligent computing: theory and applications. Springer, Berlin, pp 611–619
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Howe FA, Ye X (2017) Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int J Comput Assist Radiol Surg 12:183–203
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Szenkovits A, Meszlényi R, Buza K, Gaskó N, Lung RI, Suciu M (2018) Feature selection with a genetic algorithm for classification of brain imaging data. Advances in feature selection for data and pattern recognition. Springer, Berlin, pp 185–202
Tampu IE, Haj-Hosseini N, Eklund A (2020) Does contextual information improve 3D U-Net based brain tumor segmentation? arXiv:2010.13460
Uribe VM (1986) Psychiatric symptoms and brain tumor. Am Fam Physician 34:95–98
Vidyarthi A, Mittal N (2017) Texture based feature extraction method for classification of brain tumor MRI. J Intell Fuzzy Syst 32:2807–2818
Vishnuvarthanan A, Rajasekaran MP, Govindaraj V, Zhang Y, Thiyagarajan A (2017) An automated hybrid approach using clustering and nature inspired optimization technique for improved tumor and tissue segmentation in magnetic resonance brain images. Appl Soft Comput 57:399–426
Wang S, Chen M, Li Y, Zhang Y, Han L, Wu J, Du S (2015) Detection of dendritic spines using wavelet-based conditional symmetric analysis and regularized morphological shared-weight neural networks. Comput Math Methods Med. https://doi.org/10.1155/2015/454076
Weninger L, Liu Q, Merhof D (2019) Multi-task learning for brain tumor segmentation. International MICCAI brainlesion workshop. Springer, Berlin, pp 327–337
Wong KC, Syeda-Mahmood T, Moradi M (2018) Building medical image classifiers with very limited data using segmentation networks. Med Image Anal 49:105–116
Xiaozhou Y (2020) Linear discriminant analysis, explained. https://towardsdatascience.com/linear-discriminantanalysis-explained-f88be6c1e00b. Accessed 1 Mar 2021
Xue Y, Yang Y, Farhat FG, Shih FY, Boukrina O, Barrett A, Binder JR, Graves WW, Roshan UW (2019) Brain tumor classification with tumor segmentations and a dual path residual convolutional neural network from MRI and pathology images. International MICCAI brainlesion workshop. Springer, Berlin, pp 360–367
Zhang Y, Wang S (2015) Detection of Alzheimer’s disease by displacement field and machine learning. PeerJ 3:e1251
Zhuge Y, Ning H, Mathen P, Cheng JY, Krauze AV, Camphausen K, Miller RWJMP (2020) Automated glioma grading on conventional MRI images using deep convolutional neural networks. Med Phys. https://doi.org/10.1002/mp.14168
Funding
There is no funding received for this research.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this research.
Corresponding author
Ethics declarations
Conflict of interest
All authors declared that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharif, M.I., Li, J.P., Amin, J. et al. An improved framework for brain tumor analysis using MRI based on YOLOv2 and convolutional neural network. Complex Intell. Syst. 7, 2023–2036 (2021). https://doi.org/10.1007/s40747-021-00310-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00310-3