Abstract
Background
The interaction between genes and the environment in psoriasis is firmly coupled by epigenetic modification. Epigenetic modifications are inherited variations in gene expression devoid of DNA sequence alterations. Non-coding RNAs are regarded as one of the epigenetic modifications that lead eventually to enduring heritable variations in gene expression. In the present study, we chose the lncRNA, Psoriasis-susceptibility-Related RNA Gene Induced by Stress (PRINS) known to have a regulatory role in psoriasis and deduced its axis of lncRNA-miRNA-mRNA through an in silico data analysis. We aimed to assess the expression levels of this lncRNA-miRNA-mRNA in patients with psoriasis to elucidate their possible roles in psoriasis management.
Methods
We investigated the lncRNA-PRINS and its target microRNAs (miRNA124-3p, miRNA203a-5p, miRNA129-5p, miRNA146a-5p, miRNA9-5p) and partner genes (NPM, G1P3) expression levels in the plasma of 120 patients with psoriasis compared to 120 healthy volunteers using quantitative real-time polymerase chain reaction and correlated the results with the patients’ clinicopathological data. Finally, we performed a function, disease, and pathway enrichment analysis for the LncRNA-miRNA-mRNA axis under study.
Results
The lncRNA PRINS, G1P3, and NPM genes showed significantly under-expressed levels while all miRNAs included in the study showed significant over-expression in patients with psoriasis relative to controls. The lncRNA PRINS, G1P3, and NPM genes showed a significant direct correlation with each other and inverse significant correlations with all miRNAs under study. All the study biomarkers showed significant results for discriminating between patients with psoriasis and controls using a receiver operating curve analysis with sensitivity over 90% except for PRINS, which was 74.2%. The G1P3 gene showed a direct significant correlation with body mass index in patients with psoriasis (p = 0.009) and an inverse significant correlation with age (p = 0.034). The NPM gene showed a significant correlation with body mass index in patients with psoriasis (p = 0.002).
Conclusions
Based on our results, we suggest that restoring the altered PRINS-miRNA-mRNA axis gene expression levels might represent a tool to prevent psoriasis worsening, along with standard therapy. Thus, on the clinical practice level, the PRINS-miRNA-mRNA axis expression profile can be utilized in designing specific targeted therapy aimed at applying a personalized medicine approach among patients with psoriasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The lncRNA PRINS, G1P3, and NPM genes showed significantly under-expressed levels, while all target microRNAs (miRNA124-3p, miRNA203a-5p, miRNA129-5p, miRNA146a-5p, miRNA9-5p) showed significant over-expression in patients with psoriasis relative to controls. |
The lncRNA PRINS, G1P3, and NPM genes showed a significant direct correlation with each other and inverse significant correlations with all microRNAs under study. |
All the study biomarkers showed significant results for discriminating between patients with psoriasis and controls using a receiver operating curve analysis with sensitivity over 90% except for PRINS, which was 74.2%. |
Our results suggest that restoring the altered PRINS-miRNA-mRNA axis gene expression levels might represent a tool to prevent psoriasis worsening, along with standard therapy. |
Based on our results, on the clinical practice level, the PRINS-miRNA-mRNA axis expression profile can be utilized in designing specific targeted therapy aimed at applying a personalized medicine approach among patients with psoriasis. |
1 Introduction
Psoriasis is an inflammatory skin disease with a multifactorial origin that causes significant stress and morbidity. It presents often with well-demarcated, scaling, and erythematous plaques at the extensor surfaces of the knees and elbows. The prevalence varies between 0.7 and 2.9% worldwide [1] and is estimated to be 0.19% among the Egyptian population [2], varying among different ethnic populations and geographical regions [3] with a preference for the Caucasian population.
Psoriasis is a complex polygenic disease in which exposure of hereditarily predisposed persons to specific environmental stressors could provoke disease development [4]. The interaction between genes and the environment in psoriasis is firmly coupled by epigenetic modification [4]. Epigenetic modifications are inherited variations in gene expression devoid of DNA sequence alterations [5]. DNA methylation, histone modification, and non-coding RNAs are regarded as epigenetic motivators, leading eventually to enduring heritable variations in gene expression [5,6,7,8]. Non-coding RNAs are RNA molecules not translated to functional proteins [9]. Evidence implied that long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) play substantial roles in epigenetic control [9]. Long non-coding RNAs are transcripts longer than 200 bases that take part in regulating chromatin dynamics, gene expression, growth, differentiation, and development [10]. The lncRNA PRINS (Psoriasis-susceptibility-Related RNA Gene Induced by Stress) is a primate conserved transcript, highly expressed in the epidermis of non-lesional psoriatic skin [11]. Elevated levels of PRINS were suggested to contribute to psoriasis pathogenesis by adjusting the response of keratinocytes to stress [11].
The interferon (IFN) inducible (G1P3) gene and the nucleophosmin (NPM) were identified as a target gene and an interacting partner protein of PRINS, respectively [12, 13]. Down-expression of PRINS in ultraviolet-irradiated keratinocytes was associated with altered expression of G1P3, and re-allocation of NPM from the nucleus to the cytoplasm led to diminished keratinocyte sensitivity for spontaneous apoptosis [11]. MicroRNAs are highly conserved RNA molecules, with an average length of 22–25 bases. The bind to the 3′-untranslated region of target messenger RNAs (mRNAs), and according to the complementary level, they either inhibit mRNA translation or accelerate its turnover, ultimately suppressing the expression of protein [14]. Data suggested that different miRNAs may take part in the pathogenesis of psoriasis. They might be involved in the regulation of hyperproliferation and differentiation of keratinocytes in addition to the abnormal immune activation noted in psoriasis [15].
In the present study, we hypothesized that our chosen RNAs by an in silico data analysis might be potential biomarkers for psoriasis. We chose a lncRNA, PRINS known to have a regulatory role in psoriasis and linked it, at the epigenetic level, with its target miRNAs and mRNAs to create a postulated lncRNA PRINS-miRNA-mRNA axis to be used as a circulating biomarker panel in psoriasis. This axis encompassed the lncRNA PRINS and its target miRNAs (miRNA124, miRNA203a, miRNA129, miRNA146a, miRNA9) and partner genes (NPM, G1P3). The aforementioned biomarker gene expression profile was investigated in the plasma of patients with psoriasis compared to healthy volunteers to elucidate their possible roles in psoriasis management.
2 Subjects and Methods
2.1 Study Participants
The present study included 240 participants. The study subjects were divided into two groups. (1) Study Group: 120 Egyptian patients diagnosed with chronic plaque psoriasis of both sexes with age above 16 years were recruited from the Dermatology Outpatient Clinics, Suez Canal University Hospital, Ismailia, Egypt and we excluded patients with any other autoimmune diseases. (2) Control Group: 120 healthy non-related participants of Egyptian descent, matched by age and sex to the study group with no family history of psoriasis or any autoimmune disease. The clinicopathological data, including patients’ age, sex, body mass index (BMI), family history, age of disease onset, disease duration, severity, and lines of treatment were collected from the patients’ history. All patients were subjected to a detailed dermatological examination to determine the following: the site, size, pattern, and distribution of individual lesions and an assessment of disease severity according to the Psoriasis Area and Severity Index (PASI) score, which included an assessment of four body areas: head and neck, upper limbs, trunk, and lower limbs. Within each area, the severity of three signs, erythema, thickness/induration, and desquamation/scaling, is each assessed on a five-point scale: 0, none; 1, mild; 2, moderate; 3, severe; 4, very severe. According to the European consensus, a PASI score of < 10 is mild, 10–20 is moderate, and < 20 is severe [16].
2.2 Selection of LncRNA-miRNA-mRNA Under Study Using Bioinformatics Tools
The LncRNA-miRNA-mRNA genes under study were selected using the bioinformatics online tools, HMDD (http://www.cuilab.cn/) [17], miR2Disease (http://www.mir2disease.org/) [18], LncRNADisease version 2.0 (http://www.rnanut.net/lncrnadisease/index.php/home) [19].
2.3 Sample Collection and Total RNA Extraction Including miRNA
Three milliliters of fresh venous blood was collected in vacutainer tubes containing ethylenediaminetetraacetic acid anticoagulant. Blood samples were centrifuged to separate plasma, 100 μL of plasma was then preserved in a 500-μL Qiazole reagent. The plasma samples were stored at − 80 °C until further analysis.
Total RNA, including miRNA, was extracted from plasma using the Qiagen (Germantown, Maryland, USA) miRNeasy mini kit (Qiagen, Catalog no. 217004) following the protocol supplied by the manufacturer. An Eppendorf 5417C cooling microcentrifuge with adjusted temperature was used throughout the RNA extraction process. RNA concentration and purity were determined using the NanoDrop 2000 1C spectrophotometer at the absorbance of 260 and 280 nm (NanoDrop Tech., Inc., Wilmington, DE, USA). A ratio between 1.8 and 2.2 was considered acceptable for further genetic analysis.
2.4 LncRNA-miRNA-mRNA Gene Expression Analysis
The total RNA extracted was subjected to reverse transcription where complementary DNA was generated from total RNA containing miRNA with the miScript II RT Kit (Qiagen, Catalog no. 218161), in which miRNAs and other noncoding RNAs are polyadenylated by poly(A) polymerase and converted into complementary DNA by reverse transcription with oligo-dT priming. Reverse transcription was carried out in a Veriti™ 96-Well Thermal Cycler (Applied Biosystems, Waltham, MA, USA) at 37 °C for 1 h, followed by inactivation of the reaction by briefly incubating at 95 °C.
LncRNA-miRNA-mRNA expression profiling was carried out using real-time polymerase chain reaction (PCR) based on SYBR Green. The premix of complementary DNA was used as a template for real-time PCR relative quantification of lncRNA, miRNA, and mRNA expression. Primers for the LncRNA PRINS, miRNAs (miRNA124-3p, miRNA203a-5p, miRNA129-5p, miRNA146a-5p, miRNA9-5p), and (NPM, G1P3) mRNA are described in Table 1 and the miScript SYBR Green PCR Kit (Qiagen, cat. no 218076) was used to measure the expression levels. GAPDH and SNORD68 were used as endogenous controls to enable a data analysis using the ΔΔCT method of relative quantification. The expression levels were measured according to the quantitative real-time PCR experiments minimal information required for publication guidelines. “No-template” and “no-reverse transcribed” controls were included in each run and all reactions run in duplicate. The PCR run initially at 95 °C for 5 min, followed by 40 cycles at 95 °C (15 s), then at 55 °C (1 min), and finally at 72 °C (1 min) for denaturation, annealing, and elongation, respectively.
2.5 Gene Expression Data Analysis
Fold changes for all the circulating study biomarkers in each psoriasis sample relative to its control were estimated using the Livak method [20] based on the (threshold) cycle (Cq or CT) quantification value as follows: relative expression = 2−ΔΔCq, where ΔΔCq = (Cq Cir-Biomarker − Cq SNORD68/GAPDH)psoriasis − (Cq Cir-Biomarker − Cq SNORD68/GAPDH)control.
2.6 Function, Disease, and Pathway Enrichment Analysis of the LncRNA-miRNA-mRNA Under Study
Function, disease, and pathway enrichment of the LncRNA-miRNA-mRNA under study were analyzed using gene analytics software (https://geneanalytics.genecards.org) [21]. The score for disease analysis was based on the following: (a) the number of biomarkers under study matching psoriasis normalized by the whole gene numbers specifically associated with psoriasis. (b) The type and quality of the disease–gene relationship. These relationships are based on MalaCards sources including: (1) differentially expressed genes found to be significantly over-expressed or under-expressed in psoriasis in comparison to normal controls. The differentially expressed genes were calculated by an algorithm for filtering genes whose expressions are significantly variable among the psoriasis samples. Then, using the e-Bayesian method, differentially expressed genes were identified. Any gene was defined as differentially expressed if it is over-expressed or under-expressed by more than twofold, and the p value is equal to 0.05 or lower. (2) Genetic Association Sprites were determined from several sources available on MalaCards for genetic associations with psoriasis. For each gene, the maximal score was used as the final biomarker score.
For pathways and gene ontology terms that match our biomarkers under study, the matches were presented in the order of the matching scores. The null hypothesis was tested using a binomial distribution test that the input biomarkers were not highly represented within any pathway or gene ontology term. The score shown for each biomarker was the resulting p value, the higher the score, the better the matching. The maximum score was assigned for results with p values lower than 10–50.
2.7 Statistical Analysis
We used to Statistical Package for the Social Sciences (SPSS) for Windows software version 26.0 (IBM Corporation, Armonk, NY, USA). The study power and sample size were calculated by G*Power version 3.1.9.2. The estimated power for the gene expression study design was 95% at a total sample size of 200, an alpha error probability of 0.05, and a calculated effect size of 0.5. For continuous variables, means ± standard deviations were used, while for categorical variables we estimated the frequencies and percentages. Data were tested for normality and outliers. Mann–Whitney and Student t tests were used when appropriate, to compare between psoriasis cases and controls. We used the Spearman correlation test to determine the correlation coefficient. We performed a two-sided p value correlation analysis by the Spearman’s rank test. A two-tailed p value of < 0.05 was considered statistically significant. The area under the curve (AUC) of the receiver operating characteristic (ROC) was plotted to evaluate the diagnostic and prognostic value of putative biomarkers.
2.8 Ethics Statement
The study was approved by the Suez Canal University, Faculty of Medicine, Ethics Committee in Ismailia, Egypt (Approval No. 4503) and conducted according to the Declaration of Helsinki’s guidelines. Informed consent was obtained from all individual participants included in the study.
3 Results
3.1 Baseline Characteristics Among the Study Population
Table 2 shows the baseline characteristics among our study population. The age distribution among the study population ranged from 16.0 to 60 years, where the mean age for the patients was 41.35 ± 14.16 years and the mean age for the controls was 37.6 ± 11.83 years, with no statistically significant difference between both groups. Regarding special habits, 66.6% of patients were non-smokers and 33.3% were smokers while 62.5% of controls were non-smokers and 37.5% were smokers. Concerning BMI (kg/m2), the mean BMI was 26.60 ± 3.91 kg/m2 for patients while it was 27.70 ± 3.93 kg/m2 for controls with no statistical difference among both groups and different obesity grades. The mean age of disease onset was 34.71 ± 13.68 years and the mean duration was 6.83 ± 6.24 years. According to the age of disease onset, the patients were classified into three subgroups: (1) very early-onset psoriasis: up to 20 years of age (28 patients); (2) middle early-onset psoriasis: between 21 and 40 years of age (48 patients); and (3) late-onset psoriasis: above 40 years of age (44 patients). Forty-four percent of patients showed mild severity, 34.2% showed moderate severity, and 21.7% was severe. The distribution of the studied cases according to the PASI score ranged between 2.50 and 42.50 and the mean was 12.45 ± 8.66.
3.2 Expression Signature of the Circulating lncRNA-miRNA-mRNA Under Study in Patients with Psoriasis
As depicted in Fig. 1A, B–I, the lncRNA PRINS showed significantly lower expression levels in patients with psoriasis relative to controls: − 3.5 (− 9.3 to 1.1), p < 0.001. Additionally, G1P3 and NPM genes showed significant lower expression levels in patients with psoriasis relative to controls: − 6.9 (− 9.5 to − 3.5), p < 0.001 and − 9.8 (− 13.2 to − 6.5), p < 0.001, respectively. However, all miRNAs included in the study showed significant over-expression levels in patients with psoriasis relative to controls; miRNA146a [9.9 (7.2–13.6)], miRNA129 [9.5 (7.8–12.2)], miRNA124 [5.5 (3.3–7.5)], miRNA203a [7.1 (4.4–9.9)], and miRNA9 [7.6 (5.2–10.6)] with p < 0.001.
Circulating lncRNA-miRNA-mRNA under study expression levels. A Heat map representing the Log2 fold change for all the study biomarkers (PRINS, G1P3, NPM, miRNA146a, miRNA129, miRNA124, miRNA203a, miRNA9) included in the study. Graphs B–I show the differential expression levels of the genes and miRNAs under study in the plasma of patients with psoriasis compared with controls. Values are presented as median (Q1–Q3). For the p value calculation, a Mann–Whitney U test was applied
3.3 Circulating lncRNA-miRNA-mRNA Under Study Predictive Significance by ROC Curve Analysis
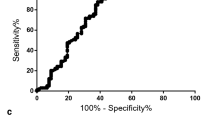
All the study biomarkers showed significant results for discriminating between patients with psoriasis and controls using the ROC curve as shown in Fig. 2 from the A–H plots. Plasma PRINS levels can discriminate between patients with psoriasis and controls with 74.2% sensitivity and 100% specificity (AUC was 0.742 at the cut-off point of 1.065-fold, p < 0.001). Plasma G1P3 levels can discriminate between patients with psoriasis and controls with 98.3% sensitivity and 100% specificity (AUC was 0.983 at the cut-off point of 1.75-fold, p < 0.001). Plasma NPM levels can discriminate between patients with psoriasis and controls with 99.2% sensitivity and 100% specificity (AUC was 0.992 at the cut-off point of 2.33-fold, p < 0.001). Plasma miRNA146a and 129 levels can discriminate between patients with psoriasis and controls with 100% sensitivity and 100% specificity (AUC was 1, p < 0.001). Plasma miRNA124 levels can discriminate between patients with psoriasis and controls with 88.3% sensitivity and 100% specificity (AUC was 0.883 at the cut-off point of 0.895-fold, p < 0.001). Plasma miRNA203a levels can discriminate between patients with psoriasis and controls with 94.2% sensitivity and 100% specificity (AUC was 0.942 at the cut-off point of 0.765-fold, p < 0.001). Plasma miRNA9 levels can discriminate between patients with psoriasis and controls with 96.7% sensitivity and 100% specificity (AUC was 0.967 at the cut-off point of 0.88-fold, p < 0.001).
Circulating lncRNA-miRNA-mRNA under study predictive significance by receiver operating characteristic (ROC) curve analysis. Graphs A–H show the ROC curve including area under the curve (AUC) ± standard error (SE) with upper and lower asymptomatic 95% confidence interval (CI) value for each biomarker
3.4 Correlation Analysis of the Circulating lncRNA-miRNA-mRNA Under Study and the Clinicopathological Features of Patients with Psoriasis
The correlation of all the study circulating biomarkers with each other is shown in Fig. 3. The lncRNA PRINS, G1P3, and NPM genes showed a significant direct correlation with each other and inverse significant correlations with all miRNAs under study. For all miRNAs under study, miRNA146a, miRNA129, miRNA124, miRNA203a, and miRNA9 showed inverse significant correlations with PRINS, G1P3, and NPM and direct significant correlations with each other.
The correlation of all the study circulating biomarkers with the clinicopathological features of patients with psoriasis is shown in Table 3. The G1P3 gene showed a direct significant correlation with BMI in patients with psoriasis (p = 0.009, r = 0.169) and an inverse significant correlation with age (p = 0.034, r = − 0.137). The NPM gene showed a significant correlation with BMI in patients with psoriasis (p = 0.002, r = 0.198).
A correlational analysis between the clinicopathological characteristics revealed a direct significant correlation between PASI score and disease severity (p < 0.001, r = 0.912), disease duration (p = 0.004, r = 0.262), and BMI (p = 0.045, r = 0.184). The age of onset of disease showed a direct significant correlation with BMI (p = 0.037, r = 0.191), family history (p < 0.001, r = 0.335), and the patient’s age (p < 0.001, r = 0.896). Finally, the age of the patient showed a direct correlation with PASI (p = 0.000, r = 0.335), severity (p = 0.035, r = 0.193), BMI (p < 0.001, r = 0.0429), disease duration (p = 0.003, r = 0.272) and disease onset (p < 0.001, r = 0.896) and an inverse correlation with family history (p = 0.001, r = −0.293).
3.5 Pathway, Disease, and Function Enrichment Analysis of the Circulating lncRNA-miRNA-mRNA Under Study
The pathway matching score shown in Fig. 4A showed the top ten pathways related to our studied biomarkers as: Alzheimer’s disease pathway, cell differentiation index, BARD1 signaling events, melatonin metabolism and effects, crizotinib pathway, Aurora B signaling, transcriptional regulation by the AP-2 family of transcription factors, TP53 regulates transcription of cell cycle genes, miRNAs involved in DNA damage response, and the HIF-1-α transcription factor network. Considering the disease matching score results shown in Fig. 4B, psoriasis was ranked in the fourth position in the disease list that can be caused by the circulating LncRNA-miRNA-mRNA under study with a score value of 4.12. These results were based on MalaCards sources and included genetic associations and differentially expressed genes in psoriasis in comparison to normal controls.
The gene ontology analysis of all our study biomarkers is presented in Fig. 5, where the cellular component matching score revealed the highest ten scores were in the granular component, spindle pole centrosome, large ribosomal subunit, nuclear speck, small ribosomal subunit, protein-DNA complex, extracellular exosome, ribonucleoprotein complex, nucleolus, and microtubule organizing center, as shown in Fig. 5A. The top ten molecular functions for the circulating LncRNA-miRNA-mRNA under study were as follows: mRNA binding involved in post-transcriptional gene silencing, mRNA 3′-untranslated region binding, high-density lipoprotein particle binding, Tat protein binding, ribosomal large subunit binding, ribosomal small subunit binding, transcription factor binding, core promoter sequence-specific DNA binding, NF-κB binding, and DNA-binding transcription factor binding, as presented in Fig. 5B. Finally, considering the top ten biological processes for our circulating LncRNA-miRNA-mRNA under study, Fig. 5C shows their order according to their matched score as follows: negative regulation of cytokine production involved in the inflammatory response, negative regulation of interleukin-8 production, gene silencing by miRNA, negative regulation of gene expression, negative regulation of xenobiotic detoxification by transmembrane export across the plasma membrane, miRNA-mediated inhibition of translation, negative regulation of transporter activity, negative regulation of NIK/NF-κB signaling, positive regulation of apoptotic process in bone marrow cells, and regulation of mRNA stability involved in the cellular response to ultraviolet.
4 Discussion and Conclusions
In recent years, transcriptome studies have uncovered non-coding RNAs as crucial players in psoriasis through altering keratinocyte functions and the dysregulation of inflammatory reactions in affected skin [22, 23]. In the present study, we investigated the expression of LncRNA PRINS, its target miRNAs, and mRNAs in patients with psoriasis. The expression of PRINS was significantly down-regulated in patients with psoriasis. Previous studies demonstrated that PRINS expression is higher in both non-lesional and lesional epidermis compared with healthy epidermis, with the highest expression found in non-lesional epidermis [12, 24]
The discrepancy in PRINS expression between the present study and the previous studies could be attributed to different specimens. In the previous studies [12, 24], PRINS expression was assessed in isolated epidermis while in the present study PRINS expression was detected in plasma.
PRINS is expressed in various healthy human tissues, for example, veins, gonads, lungs, gastrointestinal tract, and skin [12], thus plasma PRINS can originate from all the producing tissues in the body. Therefore, down-regulation of PRINS in the plasma of patients with psoriasis can be a reflection for decreased PRINS, particularly in the venous epithelium and affected skin, an assumption that needs further investigation.
The down-regulation of PRINS in patients with psoriasis compared with controls could be explained as follows: the inflammatory cytokines, interleukin (IL)-17, IFN-γ, tumor necrosis factor (TNF), and IL-22 produced by infiltrating T helper-22 and T helper-1 lymphocytes at the dermal-epidermal junction [25], affect both keratinocytes and the endothelium. Interferon-γ mediates keratinocyte proliferation and promotes inflammatory responses in endothelial cells [26], by activating several signaling pathways including the signal transducer and activator of the transcription 3 (STAT3) pathway [27, 28].
STAT3 interacts with histone deacetylase 1, a key epigenetic regulator involved in re-programming gene transcription [29]. The transcription start site of the PRINS gene is marked by a high density of histone modification sites associated with active transcription, such as acetylated lysine 9 and 27 (H3K9Ac, H3K27Ac) [11]. Therefore, the de-acetylation of lysine residues mediated by histone deacetylase 1 creates a more condensed chromatin structure, which could repress PRINS transcription [30]. In favor of this explanation, Sonkoly et al. stated that the treatment of non-lesional epidermal keratinocytes with a mixture of T-cell lymphokines including IFN-γ induces both keratinocyte hyperproliferation and a significant reduction in PRINS expression [24].
In addition, STAT3 activity is enhanced in skin affected by psoriasis and is responsible for the hyperproliferation and altered differentiation of the psoriatic epidermis [31]. Moreover, the expression of histone deacetylase 1 was observed to increase significantly in psoriatic lesions and peripheral blood mononuclear cells of patients with psoriasis compared with healthy controls [32, 33].
PRINS was proposed to contribute to psoriasis pathogenesis by altering the stress response of non-lesional epidermis and keratinocyte innate immune responses [11]. Danis et al. suggest a broad anti-inflammatory function for PRINS, providing insight into the protective role of high PRINS expression in psoriatic non-lesional skin [34]. However, the exact mechanisms are not yet clear. It was proposed that the nuclear and cytoplasmic localization of PRINS could contribute to psoriasis by modifying the expression of proteins and miRNAs, which have roles in psoriasis pathogenesis [11].
G1P3 genes and NPM were under the control of and the direct interacting partner of PRINS, respectively [35]. G1P3 and NPM had been previously shown to play important roles in psoriasis pathogenesis [36].
In the present study, the down-regulated expression of PRINS was associated with the down-regulated expression of both G1P3 and NPM in patients compared with controls. A result that agrees with a previous study that found that the suppression of PRINS resulted in down-regulated G1P3 mRNA expression [35]. Conversely, our result disagrees with the previous that silencing of PRINS resulted in a slight elevation of NPM expression [12], which could be attributed to different methodologies in both studies. Generally, lncRNAs mediate transcriptional regulation by several mechanisms. They modulate the chromatin state at a target gene locus by interacting with and recruiting chromatin-modifying enzymes, which leads to the activation of gene transcription. In addition, they interact with other RNA-binding factors to form RNA–protein complexes that recruit key proteins to the target gene promoters, promoting transcription [37].
Regarding the correlation between the clinicopathological features and the biomarkers under study, G1P3 expression showed a direct significant correlation with BMI in patients with psoriasis. G1P3 is an IFN-induced gene, particularly IFNγ, which is up-regulated in direct proportion to the increase in BMI [38]. Conversely, NPM expression showed a negative correlation with BMI. NPM regulates the levels of reactive oxygen species (ROS) at least partially via regulating the expression of peroxiredoxin-6, a member of the peroxiredoxin family of antioxidant enzymes, widely expressed in all tissues [39]. Liu et al. reported that ROS was increased after NPM knockdown due to down-regulated peroxiredoxin-6 [40]. High levels of ROS are linked to obesity [41], as well as peroxiredoxin-6, which is linked to obesity by regulating mitochondrial oxidative stress and ROS production [42].
However, the expression of miRNA124, miRNA203a, miRNA129, miRNA146a, and miRNA9 were up-regulated, in an inverse correlation with PRINS expression validating the in-silico data analysis that those miRNAs are possible targets of PRINS. PRINS by interacting with complementary sequences in miRNAs can function as a “sponge,” which could constrain miRNA availability and activity [43]. Accordingly, the down-expression of PRINS resulted in the release of those miRNAs, which could explain their up-regulated expression in the present study.
The high expressions of miRNA146a, miRNA203a, and miRNA124 in psoriasis lesions agree with previous research [44,45,46]. Up-regulated expression of miRNA146a in both lesions and peripheral blood mononuclear cells of patients with psoriasis was positively correlated with IL-17 expression. As mentioned, IL-17 may contribute to the hyperproliferation of keratinocytes because of persistent inflammation in psoriatic lesions [44].
Enhanced expression of miRNA203a in psoriasis-affected keratinocytes was associated with down-regulation of the suppressor of cytokine signaling-3 (SOCS-3), a negative regulator of the STAT3 pathway [47, 48]. Sustained activation of STAT3 signaling up-regulates the transcription of TGF-β, EGFR, and IL-6 genes, blocks apoptosis, favors cell proliferation and survival, and promotes angiogenesis [49].
MiRNA124 is another miRNA that might affect human keratinocyte proliferation and migration and impact the inflammatory microenvironment in psoriatic lesions through modulating IL-17A and TNF expression [45]. MiRNA124 negatively regulates several pathways including STAT3 and TLRs [50]. Up-regulation of the miRNA124 target STAT3 pathway led to decreased IL-6 production and TNF-α release owing to reduced activity of the TNF-α-converting enzyme [51]. Add-on miRNA124 targets multiple components of the TLR signaling, including TLR6, TNF-α, and TNF receptor-associated factor 6, collectively resulting in the prevention of excessive inflammation [52].
It is worth noting that, to our knowledge, this is the first study that documents an up-regulation of miRNA129 and miRNA9 in patients with psoriasis. Overexpression of miRNA129 significantly promoted CD3+/CD4+ T-lymphocyte proliferation, inhibited T-cell apoptosis, and increased IL-2 and IFN expression levels through targeting SOXS6 [53]. T helper-17, a subset of CD4+ T-cell-derived proinflammatory cytokines including IL-17A, IL-17F, IL-21, IL-22, and IL-26, plays a critical role in the pathogenesis of psoriasis [54]. MiRNA9 is another miRNA that has an anti-inflammatory effect. Overexpression of miRNA9 is correlated with reduced NF-κB signaling pathway-related factors such as NF-κB, p65, TNF-α, IL-1β, and IL-6 [55]. As a result, it can be assumed that down-regulation of PRINS and associated up-regulation of both proinflammatory and anti-inflammatory miRNAs may prevent excessive inflammatory reactions and contribute to the low-grade inflammation and chronicity characteristic of psoriasis [56].
Gene ontology of the PRINS lncRNA-miRNA-mRNA under study revealed that the axis is functioning mainly in the regulation of cytokine production involved in the inflammatory response, gene silencing by miRNA, and regulation of gene expression, data in line with previous studies [37, 44,45,46,47,48,49,50,51,52,53,54,55]. The biological functions are correlated with the molecular function as the LncRNA-miRNA-mRNA under study are involved in gene silencing at both the transcriptional and post-transcriptional levels by binding to transcription factor, TAT protein (a regulator of gene expression), mRNA 3′-untranslated region, and large and small ribosomal subunits [14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58].
The PRINS LncRNA-miRNA-mRNA axis components are mainly active in the microtubule organizing center, nucleolus, ribonucleoprotein complex, and extracellular exosome. These organelles contribute to the process of cellular differentiation, cell division, and organization [58].
The pathway matching score and the disease matching score results showed that the circulating lncRNA-miRNA-mRNA under study are related to different pathways affecting cell proliferation, differentiation, and apoptosis [59,60,61], and in addition to psoriasis, are associated with different types of cancer especially hepatocellular carcinoma and prostate and gastric cancer [62,63,64]. These findings suggest that patients with psoriasis, having an altered gene expression profile for the biomarkers under study, could be more susceptible to other diseases mentioned in Fig. 4 that are mostly cancer or autoimmune related.
In conclusion, with the given insights of the present study and others [45, 46, 65], it could be postulated that restoring the altered PRINS and miRNAs physiological levels might represent a tool to prevent psoriasis worsening, along with standard therapy. RNA therapy, based on supplying exogenous genetically engineered mRNA to replenish the down-regulated endogenous mRNA [66] and/or miRNA activity modulators as anti-miRNA antisense oligonucleotides to reduce the up-regulated miRNA, could be delivered to target cells by encapsulation into lipid-based nanoparticles [67, 68].
The advantages of RNA-based drugs are their potential to target the different levels of gene expression, and a reduced risk of genotoxicity [66]. PRINS and its target miRNA expression profile can be utilized in designing specific targeted RNA-based therapy for every patient aiming to reduce costs, duration, and comorbidity associated with psoriasis. Thus, applying the concept of personalized medicine in psoriasis.
Considering the current study limitations that could be addressed in future research, our study has three major limitations. The first limitation is that our laboratory work was confined to investigating the biomarkers under study expression profile followed by the bioinformatics analysis for the results. We did not have the opportunity to conduct a further functional analysis. Thus, we recommend doing both in vitro and in vivo functional studies for these biomarkers in psoriasis, thus helping to provide better insights for their actual role. The second limitation is the size of the cohort under study, a larger sample size from multicentric sources is recommended in future studies for confirming the study results on a wider scale. Finally, the third limitation was the scarcity of previous research studies using these biomarkers in psoriasis, this was an opportunity to pin point this literature gap and to emphasize the need for further research in this area.
References
Becher B, Pantelyushin S. Hiding under the skin: interleukin-17–producing γδ T cells go under the skin? Nat Med. 2012;18(12):1748–50.
Parisi R, Symmons DP, Griffiths CE, Ashcroft DM. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Investig Dermatol. 2013;133(2):377–85.
Springate DA, Parisi R, Kontopantelis E, Reeves D, Griffiths CE, Ashcroft DM. Incidence, prevalence and mortality of patients with psoriasis: a UK population-based cohort study. Br J Dermatol. 2017;176(3):650–8.
Feinberg AP, Fallin MD. Epigenetics at the crossroads of genes and the environment. JAMA. 2015;314(11):1129–30.
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–3.
Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8.
Ptashne M. On the use of the word ‘epigenetic.’ Curr Biol. 2007;17(7):R233–6.
Bernstein E, Allis CD. RNA meets chromatin. Gen Dev. 2005;19(14):1635–55.
Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics. Oncol Rep. 2017;37(1):3–9.
Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–64.
Széll M, Danis J, Bata-Csörgő Z, Kemény L. PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch. 2016;468(6):935–43.
Szegedi K, Göblös A, Bacsa S, Antal M, Németh IB, Bata-Csörgő Z, et al. Expression and functional studies on the noncoding RNA. PRINS Int J Mol Sci. 2013;14(1):205–25.
Colombo E, Alcalay M, Pelicci PG. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30(23):2595–609.
Chandra A, Ray A, Senapati S, Chatterjee R. Genetic and epigenetic basis of psoriasis pathogenesis. Mol Immunol. 2015;64(2):313–23.
Løvendorf MB, Mitsui H, Zibert JR, Røpke MA, Hafner M, Dyring-Andersen B, et al. Laser capture microdissection followed by next-generation sequencing identifies disease-related microRNA s in psoriatic skin that reflect systemic micro RNA changes in psoriasis. Exp Dermatol. 2015;24(3):187–93.
Chow C, Simpson MJ, Luger TA, Chubb H, Ellis CN. Comparison of three methods for measuring psoriasis severity in clinical studies (part 1 of 2): change during therapy in psoriasis area and severity index, static Physician’s Global Assessment and lattice system Physician’s Global Assessment. J Eur Acad Dermatol Venereol. 2015;29(7):1406–14.
Huang Z, Shi J, Gao Y, Cui C, Zhang S, Li J, et al. HMDD v.30: a database for experimentally supported human microRNA–disease associations. Nucleic Acids Res. 2019;47(D1):D1013–7.
Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37(Suppl_1):D98-104.
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q, Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47(D1):D1034–7.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta C(T)) method. Methods. 2001;25(4):402–8. https://doi.org/10.1006/meth.2001.1262.
Ben-Ari Fuchs S, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, et al. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. OMICS. 2016;20(3):139–51.
Kelly BJ, Fitch JR, Hu Y, Corsmeier DJ, Zhong H, Wetzel AN, et al. Churchill: an ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol. 2015;16(1):1–4.
Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Investig Dermatol. 2014;134(7):1828–38.
Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene. PRINS J Biol Chem. 2005;280(25):24159–67.
Diani M, Altomare G, Reali E. T cell responses in psoriasis and psoriatic arthritis. Autoimmun Rev. 2015;14(4):286–92.
Mehta NN, Teague HL, Swindell WR, Baumer Y, Ward NL, Xing X, et al. IFN-γ and TNF-α synergism may provide a link between psoriasis and inflammatory atherogenesis. Sci Rep. 2017;7(1):13831.
Wolk K, Haugen HS, Xu W, Witte E, Waggie K, Anderson M, et al. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-γ are not. J Mol Med. 2009;87(5):523–36.
Chrobak I, Lenna S, Stawski L, Trojanowska M. Interferon-γ promotes vascular remodeling in human microvascular endothelial cells by upregulating endothelin (ET)-1 and transforming growth factor (TGF) β2. J Cell Physiol. 2013;228(8):1774–83. https://doi.org/10.1002/jcp.24337.PMID:23359533;PMCID:PMC4072032.
Wingelhofer B, Neubauer HA, Valent P, Han X, Constantinescu SN, Gunning PT, et al. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia. 2018;32:1713–26.
Kim D, Park G, Huuhtanen J, Ghimire B, Rajala H, Moriggl R, et al. STAT3 activation in large granular lymphocyte leukemia is associated with cytokine signaling and DNA hypermethylation. Leukemia. 2021;35(12):3430–43.
Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, et al. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med. 2005;11(1):43–9.
Hwang YJ, Na JI, Byun SY, Kwon SH, Yang SH, Lee HS, et al. Histone deacetylase 1 and Sirtuin 1 expression in psoriatic skin: a comparison between guttate and plaque psoriasis. Life. 2020;10(9):157.
Zhang P, Su Y, Zhao M, Huang W, Lu Q. Abnormal histone modifications in PBMCs from patients with psoriasis vulgaris. Eur J Dermatol. 2011;21:552–7.
Danis J, Göblös A, Bata-Csörgő Z, Kemény L, Széll M. PRINS non-coding RNA regulates nucleic acid-induced innate immune responses of human keratinocytes. Front Immunol. 2017;29(8):1053.
Szegedi K, Sonkoly E, Nagy N, Németh IB, Bata-Csörgő Z, Kemény L, et al. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19(3):269–78.
Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-α production. J Exp Med. 2005;202(1):135–43.
Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, et al. Long non-coding RNAs: mechanism of action and functional utility. Noncoding RNA Res. 2016;1(1):43–50.
Chen L, Zhou Y, Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018;15(257):25–32.
Schmidt FM, Weschenfelder J, Sander C, Minkwitz J, Thormann J, Chittka T, et al. Inflammatory cytokines in general and central obesity and modulating effects of physical activity. PLoS One. 2015;10: e0121971.
Wang X, Phelan SA, Forsman-Semb K. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278(27):25179–90.
Liu GY, Shi JX, Shi SL, Liu F, Rui G, Li X, et al. Nucleophosmin regulates intracellular oxidative stress homeostasis via antioxidant PRDX6. J Cell Biochem. 2017;118(12):4697–707.
McMurray F, Patten DA, Harper ME. Reactive oxygen species and oxidative stress in obesity: recent findings and empirical approaches. Obesity (Silver Spring). 2016;24(11):2301–10.
Zhang W, Ambati S, Della-Fera MA, Choi YH, Baile CA, Andacht TM. Leptin modulated changes in adipose tissue protein expression in OB/OB mice. Obesity. 2011;19(2):255–61.
Xia P, Fang X, Zhang ZH, Huang Q, Yan KX, Kang KF, et al. Dysregulation of miRNA146a versus IRAK1 induces IL-17 persistence in the psoriatic skin lesions. Immunol Lett. 2012;148(2):151–62.
Xiao Y, Wang H, Wang C, Zeng B, Tang X, Zhang Y, et al. miR-203 promotes HaCaT cell overproliferation through targeting LXR-α and PPAR-γ. Cell Cycle. 2020;19(15):1928–40.
Xiao Y, Wang C, Zeng B, Tang X, Zhang Y, Xiang L, et al. miR124-3p/FGFR2 axis inhibits human keratinocyte proliferation and migration and improve the inflammatory microenvironment in psoriasis. Mol Immunol. 2020;1(122):89–98.
Huang RY, Li L, Wang MJ, Chen XM, Huang QC, Lu CJ. An exploration of the role of microRNAs in psoriasis: a systematic review of the literature. Medicine. 2015;94(45):e2030.
Singhvi G, Manchanda P, Rapalli VK, Dubey SK, Gupta G, Dua K. MicroRNAs as biological regulators in skin disorders. Biomed Pharmacother. 2018;1(108):996–1004.
Raaby L, Langkilde A, Kjellerup RB, Vinter H, Khatib SH, Hjuler KF, et al. Changes in mRNA expression precede changes in micro RNA expression in lesional psoriatic skin during treatment with adalimumab. Br J Dermatol. 2015;173(2):436–47.
Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–304.
Sun Y, Li Q, Gui H, Xu DP, Yang YL, Su DF, et al. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res. 2013;23(11):1270–83.
Ma C, Li Y, Li M, Deng G, Wu X, Zeng J, et al. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol. 2014;62(1):150–8.
Zhu L, Mei M. Interference of long non-coding RNA HAGLROS inhibits the proliferation and promotes the apoptosis of ovarian cancer cells by targeting miR-26b-5p. Exp Ther Med. 2021;22(2):1–8.
Li B, Huang L, Lv P, Li X, Liu G, Chen Y, et al. The role of Th17 cells in psoriasis. Immunol Res. 2020;68(5):296–309.
Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, et al. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun. 2014;5(1):1–2.
Kjær TN, Thorsen K, Jessen N, Stenderup K, Pedersen SB. Resveratrol ameliorates imiquimod-induced psoriasis-like skin inflammation in mice. PLoS One. 2015;10(5): e0126599.
Marban C, Su T, Ferrari R, Li B, Vatakis D, et al. Genome-wide binding map of the HIV-1 Tat protein to the human genome. PLoS One. 2011;6(11): e26894.
Alzyoud E, Vedelek V, Réthi-Nagy Z, Lipinszki Z, Sinka R. Microtubule organizing centers contain testis-specific γ-TuRC proteins in spermatids of Drosophila. Front Cell Dev Biol. 2021;29(9): 727264.
Chen X, He X, Guo Y, Liu L, Li H, Tan J, et al. Glucose-dependent insulinotropic polypeptide modifies adipose plasticity and promotes beige adipogenesis of human omental adipose-derived stem cells. FASEB J. 2021;35(5):e21534 (Erratum in: FASEB J. 2021 Jul;35(7):e21659).
Jian Y, Gao Z, Sun J, et al. RNA aptamers interfering with nucleophosmin oligomerization induce apoptosis of cancer cells. Oncogene. 2009;28:4201–11.
Shao J, Ding Z, Peng J, Zhou R, Li L, Qian Q, et al. MiR-146a-5p promotes IL-1β-induced chondrocyte apoptosis through the TRAF6-mediated NF-kB pathway. Inflamm Res. 2020;69(6):619–30.
Iacona JR, Lutz CS. miR-146a-5p: expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA. 2019;10(4):e1533. https://doi.org/10.1002/wrna.1533 (Epub 2019 Mar 20. PMID: 30895717).
Cheriyath V, Leaman DW, Borden EC. Emerging roles of FAM14 family members (G1P3/ISG 6–16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. 2011;31(1):173–81.
Shi P, Chen C, Li X, Wei Z, Liu Z, Liu Y. MicroRNA-124 suppresses cell proliferation and invasion of triple negative breast cancer cells by targeting STAT3. Mol Med Rep. 2019;19(5):3667–75.
Hermann H, Runnel T, Aab A, Baurecht H, Rodriguez E, Magilnick N, et al. miR-146b probably assists miRNA-146a in the suppression of keratinocyte proliferation and inflammatory responses in psoriasis. J Investig Dermatol. 2017;137(9):1945–54.
Kim YK. RNA therapy: rich history, various applications, and unlimited prospects. Exp Mol Med. 2022;54(4):455–65.
Sempere LF, Azmi AS, Moore A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip Rev RNA. 2021;12: e1662.
Yuan D, Chen Y, Li X, Li J, Zhao Y, Shen J, et al. Long non-coding RNAs: potential biomarkers and targets for hepatocellular carcinoma therapy and diagnosis. Int J Biol Sci. 2021;17(1):220–35.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). Article processing charge (APC) was funded by the Egyptian Science, Technology & Innovation Funding Authority (STDF) via the Springer Nature - Egyptian open access agreement for supporting researchers inside Egypt.
Conflict of interest
The authors have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
The study was approved by the Suez Canal University, Faculty of Medicine, Ethics Committee in Ismailia, Egypt (Approval No. 4503) and conducted according to the Declaration of Helsinki’s guidelines.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
HYA and NZT designed the study, NZT collected the clinical data, NHS collected the patients’ samples, HYA, NHS, and LATE carried out the experiments, and HYA and LATE analyzed and interpreted the patient data. All authors discussed the results, contributed to the final manuscript, and approved it for publication.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Abdallah, H.Y., Tawfik, N.Z., Soliman, N.H. et al. The lncRNA PRINS-miRNA-mRNA Axis Gene Expression Profile as a Circulating Biomarker Panel in Psoriasis. Mol Diagn Ther 26, 451–465 (2022). https://doi.org/10.1007/s40291-022-00598-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-022-00598-y