Abstract
Background
The identification of pretherapeutic somatic BRCA variants can have considerable clinical impact given that they affect response to the new poly (ADP-ribose) polymerase (PARP)-targeted therapy. One major issue with this type of testing is the identification of splicing variants of uncertain significance (VUS) on degraded somatic messenger RNA. It is therefore important to be able to quickly characterize these splice variants.
Objective
As part of PARP inhibitor targeted therapy, we have investigated a method for the direct confirmation of potential pathogenic somatic splice variants of BRCA1 found in fixed tumor samples. Previously these VUS have commonly only been tested by in silico analysis.
Methods
Five BRCA1 variants affecting splicing were characterized from formalin-fixed, paraffin-embedded (FFPE) ovarian carcinoma tissues by next-generation sequencing (NGS). Three patient samples had already been functionally characterized and were used as controls. Total somatic RNA from samples was extracted, reverse-transcribed, and amplified with several primer pairs encompassing the target exon. The polymerase chain reaction (PCR) products were analyzed by capillary gel electrophoresis to assess possible changes in size due to splicing alterations. Finally, we confirmed our results by cloning, followed by Sanger sequencing, and analyzed the expression of the aberrant forms.
Results
Our molecular approach made it possible to visualize the splicing outcomes of three variants (c.5194-2A>G, c.5434C>G, and c.547+1G>A) already identified and present in databases and/or identified with prediction tools (ClinVar, UMD, ARUP Utah database, and Human Splice Finder splices sites prediction) and to confirm their exon skipping consequences, their expression in tumors, and thus their pathogenicity. The c.4484+5G>A variant was not found in databases and was predicted to have no impact on splicing, but was found to display altered processing in tumor tissue. This variant also had a major detrimental impact on transcriptional expression.
Conclusion
In a break from purely in silico approaches, we propose a simple and rapid pretherapeutic functional analysis of somatic BRCA1 variants potentially involved in splicing alterations. This approach will allow more ovarian cancer patients to benefit from new therapies targeting PARP.




Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23(1):41–4.
Ledermann JA. Front-line therapy of advanced ovarian cancer: new approaches. Ann Oncol. 2017;28(8):46–50.
Vetter MH, Hays JL. Use of targeted therapeutics in epithelial ovarian cancer: a review of current literature and future directions. Clin Ther. 2018;40(3):361–71.
Pujade-Lauraine E. New treatments in ovarian cancer. Ann Oncol. 2017;28(8):57–60.
Reinbolt RE, Hays JL. The role of PARP inhibitors in the treatment of gynecologic malignancies. Front Oncol. 2013;3:237.
Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21.
Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96(1):56–67.
Chandrasekaran D, Manchanda R. Germline and somatic genetic testing in ovarian cancer patients. BJOG. 2018;125:1460.
de la Hoya M, Soukarieh O, López-Perolio I, Vega A, Walker LC, van Ierland Y, et al. Combined genetic and splicing analysis of BRCA1 c.[594-2A>C; 641A>G] highlights the relevance of naturally occurring in-frame transcripts for developing disease gene variant classification algorithms. Hum Mol Genet. 2016;25(11):2256–68.
Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42(10):737–48.
Wappenschmidt B, Becker AA, Hauke J, Weber U, Engert S, Köhler J, et al. Analysis of 30 putative BRCA1 splicing mutations in hereditary breast and ovarian cancer families identifies exonic splice site mutations that escape in silico prediction. PLoS One. 2012;7(12). Disponible sur: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3519833/.
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6.
Béroud C, Letovsky SI, Braastad CD, Caputo SM, Beaudoux O, Bignon YJ, et al. BRCA Share: a collection of clinical BRCA gene variants. Hum Mutat. 2016;37(12):1318–28.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:980–5.
Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67.
Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2–3):377–94.
Gaildrat P, Krieger S, Théry J-C, Killian A, Rousselin A, Berthet P, et al. The BRCA1 c.5434C−>G (p.Pro1812Ala) variant induces a deleterious exon 23 skipping by affecting exonic splicing regulatory elements. J Med Genet. 2010;47(6):398–403.
Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562(7726):217–22.
Steffensen AY, Dandanell M, Jønson L, Ejlertsen B, Gerdes A-M, Nielsen FC, et al. Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur J Hum Genet. 2014;22(12):1362–8.
Jansen AM, van der Klift HM, Roos MA, van Eendenburg JD, Tops CM, Wijnen JT, et al. RNA analysis of cancer predisposing genes in formalin-fixed paraffin-embedded tissue determines aberrant splicing. Eur J Hum Genet. 2018;26(8):1143–50.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval and Informed Consent
The study procedures were approved by the ethics committee of the Angers medical university under ref. 2020-05.
Conflict of Interest
LMC, AB, SF, JD, AP, VV and AM have no conflicts of interest to report.
Funding
This work was supported in part by a grant from Cancéropôle Grand Ouest. Amandine Billaud is supported by a grant from Institut de Cancérologie de l’Ouest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
40291_2020_452_MOESM1_ESM.eps
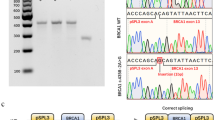
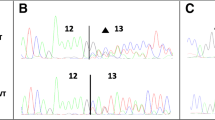
Supplementary material 1 Analysis of the c.5194-2A>G, c.4484+5G>C, c.5434C>G and c.213-1G>A variants from samples 1, 2, 3 and 5, respectively. A. The Ion Torrent data displayed in IGV from BRCA1 variant characterization. B. Sanger sequencing of the mutated BRCA1 amplicon for confirmation of the variant. C. Capillary electrophoresis analysis of the reverse transcription-PCR product framing the putative skipped exon. D. Sanger sequencing of the amplicon after cloning (EPS 3205 kb)
Rights and permissions
About this article
Cite this article
Chevalier, LM., Billaud, A., Fronteau, S. et al. Somatic mRNA Analysis of BRCA1 Splice Variants Provides a Direct Theranostic Impact on PARP Inhibitors. Mol Diagn Ther 24, 233–243 (2020). https://doi.org/10.1007/s40291-020-00452-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-020-00452-z