Abstract
Background
Mutant Phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) has been shown to be associated with the occurrence, development and prognosis in colorectal cancer (CRC). However, its detection has been limited because of complicated procedures and the low sensitivity of the present approaches.
Methods
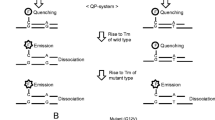
We established an ultra-sensitive peptide nucleic acid-mediated polymerase chain reaction (PNA-PCR) assay to detect PIK3CA gene mutation in exon 9 and exon 20 with cell-free DNA (cfDNA). Using this technology, we detected the mutation status of PIK3CA in 128 colorectal cancer patients. 6 CRC patients receiving targeted therapy were chosen at random to undergo continuous PIK3CA mutation detection.
Results
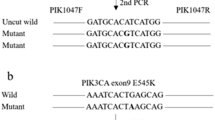
The results showed that the sensitivity of PNA-PCR clamping method was 0.1% for the exon 9 and 0.2% for the exon 20 variant alleles. When the PIK3CA mutation status was determined by PNA-PCR plus sequencing, 38.3% (49/128) of CRC carried at least one mutation, either E545Kor H1047R. The clinic-pathological parameters of age (p = 0.358), gender (p = 0.622), disease stage (p = 0.353) and disease location (p = 0.307) were not associated with the PIK3CA mutation. In the continuous monitoring study, we found that the gene status was associated with the effect of treatment. Furthermore, when the PIK3CA variant was determined by only the PNA-PCR method, there was a good linear relationship between ΔCp values and the proportion of variant DNA. The accuracy of PNA-PCR was 93.75 and 92.27% respectively when the cut-off values of ΔCp at 9.0 and 8.0 were set for determining the E545K and H1047R mutations.
Conclusions
A simple, noninvasive, ultra-sensitive PNA-PCR technology was developed and was especially suitable for the dynamic detection of PIK3CA variants using cfDNA.



Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30.
Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–7.
Wong NS, Fernando NH, Nixon AB, Cushman S, Aklilu M, Bendell JC, et al. A phase II study of capecitabine, oxaliplatin, bevacizumab and cetuximab in the treatment of metastatic colorectal cancer. Anticancer Res. 2011;31(1):255–61.
Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–5.
Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64(15):5048–50.
Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64(21):7678–81.
Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24(8):1477–80.
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554.
Wang Y, Helland A, Holm R, Kristensen GB, Borresen-Dale AL. PIK3CA mutations in advanced ovarian carcinomas. Hum Mutat. 2005;25(3):322.
Cathomas G. PIK3CA in colorectal cancer. Front Oncol. 2014;4:35.
Chong ML, Loh M, Thakkar B, Pang B, Iacopetta B, Soong R. Phosphatidylinositol-3-kinase pathway aberrations in gastric and colorectal cancer: meta-analysis, co-occurrence and ethnic variation. Int J Cancer. 2014;134(5):1232–8.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62.
He C, Zheng L, Xu Y, Liu M, Li Y, Xu J. Highly sensitive and noninvasive detection of epidermal growth factor receptor T790M mutation in non-small cell lung cancer. Clin Chim Acta Int J Clin Chem. 2013;21(425):119–24.
Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27(16):2653–9.
Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111(8):1482–9.
Yu S, Wu J, Xu S, Tan G, Liu B, Feng J. Modified PNA-PCR method: a convenient and accurate method to screen plasma KRAS mutations of cancer patients. Cancer Biol Ther. 2012;13(5):314–20.
Jeong D, Jeong Y, Park JH, Han SW, Kim SY, Kim YJ, et al. BRAF (V600E) mutation analysis in papillary thyroid carcinomas by peptide nucleic acid clamp real-time PCR. Ann Surg Oncol. 2013;20(3):759–66.
Choi JJ, Jang M, Kim J, Park H. Highly sensitive PNA array platform technology for single nucleotide mismatch discrimination. J Microbiol Biotechnol. 2010;20(2):287–93.
Hurst CD, Zuiverloon TC, Hafner C, Zwarthoff EC, Knowles MA. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res Notes. 2009;2:66.
Board RE, Thelwell NJ, Ravetto PF, Little S, Ranson M, Dive C, et al. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54(4):757–60.
Guedes JG, Veiga I, Rocha P, Pinto P, Pinto C, Pinheiro M, et al. High resolution melting analysis of KRAS, BRAF and PIK3CA in KRAS exon 2 wild-type metastatic colorectal cancer. BMC Cancer. 2013;13:169.
Li G, Luo X, He J, Zhu Z, Yu G, Qin H, et al. A novel liquidchip platform for simultaneous detection of 70 alleles of DNA somatic mutations on EGFR, KRAS, BRAF and PIK3CA from formalin-fixed and paraffin-embedded slides containing tumor tissue. Clin Chem Lab Med. 2011;49(2):191–5.
Dirican E, Kaya Z, Gullu G, Peker I, Ozmen T, Gulluoglu BM, et al. Detection of PIK3CA gene mutations with HRM analysis and association with IGFBP-5 expression levels in breast cancer. APJCP. 2014;15(21):9327–33.
Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Br Cancer Res Treat. 2010;120(2):461–7.
Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res: Official J Am Assoc Cancer Res. 2012;18(12):3462–9.
Schneck H, Blassl C, Meier-Stiegen F, Neves RP, Janni W, Fehm T, et al. Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2013;7(5):976–86.
Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2015;9(4):749–57.
Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A, Maruyama N, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Br Cancer Res Treat. 2015;150(2):299–307.
Kim ST, Lira M, Deng S, Lee S, Park YS, Lim HY, et al. PIK3CA mutation detection in metastatic biliary cancer using cell-free DNA. Oncotarget. 2015.
Emelyanova MA, Amossenko FA, Semyanikhina AV, Aliev VA, Barsukov YA, Lyubchenko LN, et al. Biochip detection of KRAS, BRAF, and PIK3CA somatic mutations in colorectal cancer patients. Mol Biol (Mosk). 2015;49(4):617–27.
Phipps AI, Ahnen DJ, Cheng I, Newcomb PA, Win AK, Burnett T. PIK3CA somatic mutation status in relation to patient and tumor factors in racial/ethnic minorities with colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2015;24(7):1046–51.
Normanno N, Rachiglio AM, Lambiase M, Martinelli E, Fenizia F, Esposito C, et al. Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann Oncol. 2015;26(8):1710–4.
He Y, Van’t Veer LJ, Mikolajewska-Hanclich I, van Velthuysen ML, Zeestraten EC, Nagtegaal ID, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res Official J Am Assoc Cancer Res. 2009;15(22):6956–62.
Liu X, Kambrick S, Fu S, Naing A, Subbiah V, Blumenschein GR, et al. Advanced malignancies treated with a combination of the VEGF inhibitor bevacizumab, anti-EGFR antibody cetuximab, and the mTOR inhibitor temsirolimus. Oncotarget. 2016.
Yen LC, Uen YH, Wu DC, Lu CY, Yu FJ, Wu IC, et al. Activating KRAS mutations and overexpression of epidermal growth factor receptor as independent predictors in metastatic colorectal cancer patients treated with cetuximab. Ann Surg. 2010;251(2):254–60.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors Qian Zeng, Li Xie, Na Zhou, Min Liu and Xianrang Song declare no conflicts of interest.
Ethical approval and informed consent
All patients declared informed consent and the study was approved by the institutional review board at the Shandong Cancer Hospital and Institute and was performed in accordance with the declaration of Helsinki.
Funding
This study was supported by Grants from the National Science Foundation of China (No. 81371886).
Rights and permissions
About this article
Cite this article
Zeng, Q., Xie, L., Zhou, N. et al. Detection of PIK3CA Mutations in Plasma DNA of Colorectal Cancer Patients by an Ultra-Sensitive PNA-Mediated PCR. Mol Diagn Ther 21, 443–451 (2017). https://doi.org/10.1007/s40291-017-0269-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-017-0269-9