Abstract
Background
New immuno-oncology (I-O) therapies that harness the immune system to fight cancer call for a re-examination of the traditional parametric techniques used to model survival from clinical trial data. More flexible approaches are needed to capture the characteristic I-O pattern of delayed treatment effects and, for a subset of patients, the plateau of long-term survival.
Objectives
Using a systematic approach to data management and analysis, the study assessed the applicability of traditional and flexible approaches and, as a test case of flexible methods, investigated the suitability of restricted cubic splines (RCS) to model progression-free survival (PFS) in I-O therapy.
Methods
The goodness of fit of each survival function was tested on data from the CheckMate 067 trial of monotherapy versus combination therapy (nivolumab/ipilimumab) in metastatic melanoma using visual inspection and statistical tests. Extrapolations were validated using long-term data for ipilimumab.
Results
Modelled PFS estimates using traditional methods did not provide a good fit to the Kaplan–Meier (K–M) curve. RCS estimates fit the K–M curves well, particularly for the plateau phase. RCS with six knots provided the best overall fit, but RCS with one knot performed best at the plateau phase and was preferred on the grounds of parsimony.
Conclusions
RCS models represent a valuable addition to the range of flexible approaches available to model survival when assessing the effectiveness and cost-effectiveness of I-O therapy. A systematic approach to data analysis is recommended to compare the suitability of different approaches for different diseases and treatment regimens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The use of traditional parametric survival functions can underestimate survival with immuno-oncology (I-O) therapies, primarily when a plateau of long term survival is observed, and therefore give a misleading estimate of life expectancy. |
Flexible models including restricted cubic splines (RCS) can provide a good fit to trial data and valid extrapolations of clinical trial endpoints, as demonstrated by the case study of progression free survival in I-O treatment of melanoma. |
Methods including the RCS-based approaches can be considered an option for survival analysis by health technology assessment bodies when considering effectiveness and cost-effectiveness. |
1 Introduction
New drugs under the class of immuno-oncology (I-O) compounds have the potential to provide lasting survival benefits and improve quality of life (QoL) for patients with cancer who previously had very few therapeutic options. Their novel pharmacodynamic and anticancer properties were first demonstrated in melanoma patients enrolled in clinical trials of ipilimumab, a monoclonal antibody that activates the immune system by targeting cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) [1, 2]. Ipilimumab is the first I-O agent approved for clinical use [3] and the therapy with the most long-term data [4].
Treatment response has historically been measured in oncology by tumour shrinkage using the Response Evaluation Criteria in Solid Tumors (RECIST) [5]. For I-O therapies, response after an initial increase in tumour burden (pseudo-progressionFootnote 1) or in the presence of new lesion(s) may result in the I-O effect being underestimated by RECIST. Therefore, to capture anti-tumour kinetics and evaluate survival endpoints accurately, the immune-related response criteria (irRECIST) were subsequently developed [6]. Under irRECIST, response patterns take account of changes in all lesions, not just target lesions (with new lesions not considered progressive disease per se) and the thresholds determining progression or response are higher than those specified by RECIST [5]. The criteria have not yet been universally adopted, with fewer than 100 PubMed citations (last checked 22 May 2017) since its origins in a series of expert workshops [7]. However, with increasing awareness of pseudo-progression, pembrolizumab trials have considered both immune-related and conventional criteria to assess response in advanced melanoma [8, 9].
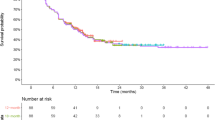
The contrasting response in I-O compared with conventional treatments is manifested in the Kaplan–Meier (K–M) curves of overall survival (OS) and progression-free survival (PFS). I-O responses have been demonstrated with ipilimumab [2], combination therapies [10] and pembrolizumab [11] in advanced melanoma and in other indications, including nivolumab in renal cell carcinoma [12]. These consistently display phases of early non-separation (between treatment and control arm), followed by separation and long-term survival (plateau) for a subgroup of patients [13]. The non-separation phase is comparable with traditional therapy and occurs within the first 3 months. The separation phase represents delayed treatment effects, where the T-cell immune response is activated, resulting in improved survival (Fig. 1a). Beyond 24 months, long-term survival occurs in a proportion of patients (in contrast with a steady decline in the comparator arm), represented by an extended plateau observed in the MDX010-20 study [2], and consistent with a pooled analysis of 10-year survival data [4].
Survival curves form the basis of estimates of life expectancy and quality-adjusted life years (QALYs) generated by economic models and used by policy makers to make reimbursement decisions on new drugs. Since a significant proportion of clinical and economic (costs and QALYs) value is reflected in the latter part (plateau) of the survival curve, it is important to understand different methods of data extrapolation to ensure that the value of I-O therapies is appropriately captured. Traditional parametric approaches, largely characterised by monotonic hazards for death from disease and used to extrapolate survival curves beyond the trial horizon, potentially underestimate the long-term impacts of I-O contained within the unique shape of the survival curve (Fig. 1b). In particular, the plateau of survival is difficult to accommodate with single parametric functions. Alternative methods that more accurately estimate the survival in I-O cohorts are therefore needed [13].
This paper evaluates the suitability of traditional parametric approaches compared with flexible models. Of these, spline-based functions are presented as a case study in the modelling and extrapolation of I-O survival data from a randomised phase III registration trial (CheckMate 067 [10]). As noted above, clinical survival endpoints, particularly OS, are critically important measures for economic evaluation. Moreover, the contrasting response patterns in I-O relative to those observed with conventional treatment [14] emphasise the importance of PFS which, in advanced cancer trials, tends to have more mature data (consistent with trial data used in this paper). As the scope for extrapolation bias is therefore less with PFS than OS [15], and long-term follow-up of I-O therapy in advanced melanoma shows similar patterns for OS and PFS [2], methods of survival analysis illustrated here were applied to PFS data. Based on the methodology adopted in this paper, recommendations are made for a systematic approach to data analysis and extrapolation for future use in economic models and I-O submissions to health technology assessment (HTA) agencies such as the National Institute for Health and Care Excellence (NICE) [16].
2 Methods
The analysis of survival functions fitted to K–M I-O survival data followed a systematic approach, initially creating internal training and validation data sets of patient-level data from CheckMate 067 (over 27 months) to provide internal validation of the survival techniques being tested. Subsequently, the heterogeneity of baseline characteristics between the data sets was assessed, K–M curves plotted and traditional and flexible survival functions fitted to the K–M data. The performance of the survival functions fitted to the data was then compared using visual inspection, statistical analysis, and assessment of consistency across data sets. Finally, the ability of the survival functions to extrapolate beyond the trial data was validated with available long-term data. Extrapolations based on a curve which provides a good fit to the data including an apparent flattening out of the K–M curve may not generate appropriate projections of survival and need to be benchmarked against longer term registry or other observational data which includes therapies with a similar mode of action [17].
2.1 CheckMate 067
CheckMate 067 (NCT01844505) was a phase III, double-blinded clinical trial of 945 treatment-naïve patients with metastatic melanoma who were randomly assigned 1:1:1 to the following regimes [10]:
-
1.3 mg/kg of nivolumab (n = 316) every 2 weeks (plus matched ipilimumab placebo)
-
2.3 mg/kg of ipilimumab (n = 315) every 3 weeks for four doses (plus matched nivolumab placebo)
-
3.1 mg/kg of nivolumab plus 3 mg/kg of ipilimumab (n = 314) every 3 weeks for four doses followed by 3 mg/kg of nivolumab every 2 weeks.
Randomisation was stratified by tumour programmed death-ligand 1 (PD-L1) status, BRAF mutation status (the gene that encodes the B-Raf protein) and American Joint Committee on Cancer metastasis stage. Treatment continued until disease progression (defined by RECIST 1.1), or when patients experienced unacceptable toxicity or withdrew from the study. A maximum treatment duration of 2 years was anticipated [18]. Patients could be treated after progression, if they had clinical benefit with no substantial adverse effects, as assessed by the investigator [10].
The analysis reported in the current paper is based on approximately 27 months of patient-level data, including a follow-up period, for the co-primary endpoint of PFS for nivolumab plus ipilimumab (n = 314) and ipilimumab (n = 315) treatment (Fig. 1a). OS data were also available; however, PFS was the preferred endpoint since the follow-up for OS was less mature, which is common in advanced cancer trials. Extrapolated data for OS are therefore more likely to be biased than those for PFS [15]. While OS may be more relevant for some decision makers, this paper focuses on the methods investigated rather than the specific data set.
2.2 Data Partitioning and Presentation
CheckMate 067 patient-level trial data were randomly partitioned using a 1:1 ratio into a ‘training’ data set and a ‘validation’ data set. The ‘training’ and ‘validation’ data sets consisted of patients with a set of common prognostic indicators and comparably defined time-to-event outcomes with similar follow-up times. Visual inspection and analytical techniques were used to evaluate the fit of a proposed model in the ‘validation’ data set. Other assessments of performance included a goodness-of-fit test and a measure of explained variation. We noted that full validation of a model requires the model to provide a complete probabilistic description of the data, sufficient to predict the survival probabilities at any relevant time point and for any combination of values of the prognostic factors.
2.3 Data Fitting Methods
The ‘training’ data set was used to assess the initial goodness of fit to the data of the survival functions tested, and the ‘validation’ data set was used to confirm the generalisability of approaches and consistency of results (as per recommended approaches for internal validation in survival analysis [15, 19]).
2.3.1 Traditional Methods
Traditional models widely used in survival analysis were selected for the first set of data analysis. Generally, they follow an underlying probability distribution with monotonic or unimodal hazards, with the general consensus that either single, multiple, or adjusted fits can achieve advanced disease risk profiles [15, 17, 20, 21]. Single fits with the common traditional parametric models (i.e. exponential, Weibull, Gompertz, log normal, and log-logistic) were applied to both treatment arms independently. Although it has been noted that traditional methods do not easily accommodate patterns of survival observed with I-O therapy, they continue to be applied in this context [22, 23] and represent the baseline against which to compare more flexible methods.
2.3.2 Flexible Methods: Combined Functions
To overcome some of the drawbacks of traditional methods, combined functions or piecewise models [17] involve the fitting of separate parametric functions to distinct phases of the K–M curve. Judgement is required to achieve an appropriate division of the data, but hazard plots, particularly the log cumulative hazard (LCH) plot, can act as a guide. Using data from the combination therapy arm as an example, LCH plots exhibited a clear change in hazard trend at 3 months (Fig. 1b). K–M up to and after the 3-month time point was therefore modelled with separate Weibull functions. As the data after 3 months did not display clear hazard trends to justify modelling of additional phases independently [15], and on grounds of parsimony, no additional models were fitted to the remaining data. Nor were other parametric functions considered. In this case, an alternative and potentially more appropriate approach to the data analysis would be to combine the K–M data for the initial 3 months with a parametric function fitted to the remainder of the K–M curve, as illustrated in Fig. 2. The function can similarly be fitted to the tail of the K–M plot if K–M data, rather than data from parametric functions, are desired for the duration of the in-trial period [20].
2.3.3 Flexible Methods: Spline-Based Models
Analogous to the combined functions approach, Royston-Parmar spline-based models are piecewise (polynomial) functions fitted sequentially to segmented portions of the data. At the border between data segments, these functions join at points known as knots [23, 24, 25] and are characterised by a high degree of smoothness at these points, lending a smooth appearance to the survival function. In contrast with the Weibull model, which imposes linearity on the relationship between LCHs and log time, restricted cubic spline (RCS) models introduce additional flexibility by allowing this relationship to be nonlinear. RCS models are constrained to be linear beyond the first and last (boundary) knots. RCS models were applied to each treatment arm independently using the stpm2 package in Stata statistical software (Fig. 3a–d) [26]. Compromise between increased flexibility and overfitting was assessed by analysing the sensitivity of the results to the number of internal knots (between one and seven). Knots were placed at evenly distributed centiles of log time for analysis [19], reducing the inherent uncertainty when seeking optimal knot placement and allowing for improved reproducibility of results. This approach mitigates the risk of overfitting, which can arise if the choice of knots or inflexion points is data driven.
Kaplan–Meier survival analysis with traditional (Weibull and log-logistic) and cubic spline (1 and 7 knots) methods for a combination therapy arm of the ‘training’ data set, b combination therapy arm of the ‘validation’ data set, c ipilimumab arm of the ‘training’ data set, and d ipilimumab arm of the ‘validation’ data set. CI confidence interval, PFS progression-free survival, RCS restricted cubic spline
2.4 Validation
2.4.1 Visual Inspection
Survival functions for the traditional and flexible models in both treatment arms were compared with the respective K–M survival curves by means of visual inspection. Interpolation was assessed independently of overall fit and based on clinical and biological plausibility, at the I-O non-separation, separation, and plateau phases (Fig. 1a). Emphasis was placed on the plateau phase given that a poor fit at the tail of the K–M plot may substantially influence extrapolation estimates. If visual inspection indicated that proportional hazards between the two treatment arms were violated, this would suggest that a single parametric function was not appropriate.
2.4.2 Statistical Methods
Akaike information criterion (AIC) and Bayesian information criterion (BIC) values for the traditional and flexible models were used to compare data fits in both treatment arms. Models with the lowest AIC and BIC values are considered to have the ‘best fit’. The generalisability of the models was also assessed. On this criterion, simple approaches were preferred to more complex models, taking account of the risk of over-parameterisation, particularly when representing the plateau.
If plots of log-cumulative hazard plots displayed linearity, traditional parametric models were deemed more suitable to estimate I-O survival. However, if plots displayed non-linearity, indicating variation in hazard patterns, flexible models were considered more appropriate [11, 12]. Additionally, goodness of fit was assessed by examining cumulative hazard plots.
2.4.3 Validation with an External Data Set
Following confirmation of acceptable consistency in results and data fits in both data sets, extrapolation of survival was conducted beyond the trial data with the traditional and flexible models and estimates compared with external PFS data reported for close to 5-year survival with ipilimumab (MDX010-20 study) [2]. Furthermore, extrapolated estimates were assessed over a 10-year horizon to assess the realism of survival trends for implementation in economic models and for use in informing policy.
All statistical analyses were performed using Stata Statistical Software (15 SE, TX: StataCorp LLC).
3 Results
3.1 Data Presentation
Baseline patient characteristics for the nivolumab plus ipilimumab (n = 314) and ipilimumab (n = 315) treatment arms were comparable across the ‘training’ and ‘validation’ data sets (Table 1). The exception was PD-L1 status, which it is thought might have a role as a biomarker in I-O (although, in advanced melanoma, evidence suggests treatment response in patients with PD-L1 positive and negative tumours [10]).
The co-primary endpoint of PFS for the ‘training’ (10.2 and 2.8 months) and ‘validation’ (13.9 and 3.1 months) data sets fell within the 95% confidence interval (CI) of the core trial data for nivolumab plus ipilimumab (11.5 months, 95% CI 8.6–16.7) and ipilimumab (2.9 months, 95% CI 2.8–3.4) (Table 1).
Visually inspecting the 95% CIs for K–M plots further supported consistency across the data sets and comparability with the core data, where CIs did not exceed the K–M estimates and CI widths were similar for all plots. Increased width was noticed in the last few observations in the combination treatment group of the ‘training’ data set, for which patient numbers were very low (Fig. 3a).
3.2 Data Fits
Visual inspection of data fits to the K–M plots on the ‘training’ data set with the traditional parametric methods (i.e. Weibull, exponential, Gompertz, log normal and log-logistic) showed a similar lack of fit in the combination therapy and ipilimumab treatment arms (Fig. 3a, c). Among the traditional methods, the log-logistic approach was the best fitting overall, distinguishing between the separation and plateau phases in the nivolumab plus ipilimumab group (n = 160), and providing the best fit to the plateau phase of the ipilimumab group (n = 149). The combined model/piecewise function did not provide a good fit at the plateau phase (Fig. 2).
RCS interpolation of the ‘training’ data set for several internal knots ranging between one and seven produced results with RCS models using various scales (proportional hazards RCS, proportional odds RCS, and probit RCS models), although the proportional hazard RCS model generally demonstrated better fits. Plots for selected proportional hazard RCS models, hereafter referred to only as RCS models, in the training data set are presented in Fig. 3a, c. For both treatment groups, aside from the early curve (non-separation and part of the separation phases) between 0 and 5 months, all RCS estimates fell within the 95% CIs and fit the K–M survival estimates well, particularly for the plateau phase. The best overall fit including the early curve (between 0 and 5 months) was seen using the RCS model with six knots. However, inspection of the plateau phase for the RCS with one internal knot revealed that this model had the best fit. Emphasis was placed on the plateau phase, as poor fit at the tail of the K–M plot would have a considerable impact on the assessment of clinical value represented by the period of extrapolation beyond the trial data.
AIC and BIC values for all data fits are presented in Table 2. Results suggest that, for both treatment arms in the ‘training’ data set, the exponential model performed least well overall. Among the traditional methods, the log normal model provided the best fit for the combination group and the Gompertz model provided the best fit for the ipilimumab group (Fig. 3a, c).
The log-cumulative hazard plots (Fig. 1b) were characterised by non-parallel lines in the first half of the plot [for ln (time) between −1 and 1], where hazard patterns (non-monotonic) differed and the treatment arms diverged, converged, and then crossed, consistent with the observed I-O phases.
For the second half of the hazard plot [for ln (time) between 1 and 3], the lines were relatively straight and parallel (monotonic), although treatment arms diverged, remained parallel, then converged. This implied that the proportional hazards assumption did not hold and a single parametric model may not be suitable to model survival. For further justification, cumulative hazard plots for each treatment were analysed for goodness of fit. Weibull, log-logistic, log normal and RCS functions with between one and seven internal knots followed the cumulative hazards closely in both treatment arms (see Fig. 4a, c for the one-knot and seven-knot RCS). For the traditional methods, this was the case for combination therapy only (but these failed to capture the ‘tail’ of the curve). For the ipilimumab arm, estimates with the traditional methods were not well matched to the cumulative hazards.
Cumulative hazard plots for a combination therapy arm of the ‘training’ data set, b combination therapy arm of the ‘validation’ data set, c ipilimumab arm of the ‘training’ data set, and d ipilimumab arm of the ‘validation’ data set. CI confidence interval, PFS progression-free survival, RCS restricted cubic spline
3.3 Validation
Visual inspection of all data fits and AIC and BIC values (Table 2) showed comparable trends in the ‘validation’ data set to those in the ‘training’ data set (Fig. 3b, d). Visual inspection revealed similar data fits overall and to distinct phases of the K–M plots (for both treatment groups, the log-logistic model of the traditional approaches and RCS models with six or seven knots were supported by AIC and BIC values). Additional analyses of cumulative hazard plots with 95% CIs for both treatment arms (Fig. 4b, d) validated selection of the RCS model with one knot (particularly for the ipilimumab arm) as the preferred estimator.
Following the confirmation of consistency in data fits across the ‘training’ and ‘validation’ data sets, extrapolation was conducted on the two data sets separately. External I-O long-term validation data were used as a proxy to compare relative trends in the slope of extrapolated survival estimates for the training and validation data sets of the ipilimumab arm, respectively (Fig. 5a, b). In the absence of long-term trial data or patient registries, this was considered a reasonable approach to validation [17].
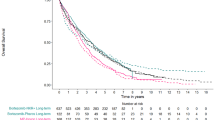
External validation with long-term data from Hodi et al. [2] (ipilimumab data) in the extrapolated estimates with traditional and cubic spline methods applied to the a ipilimumab arm of the ‘training’ data set and b ipilimumab arm of the ‘validation’ data set. PFS progression-free survival, RCS restricted cubic spline
Validation of extrapolated estimates compared with external PFS data of up to 4.6 years’ survival for ipilimumab in the MDX010-20 trial of ipilimumab plus gp100, ipilimumab alone and gp100 alone [2] showed that the RCS models provided the best fit (Fig. 5a, b). While increasing the number of knots gave a better fit overall, the one-knot model performed better than the seven-knot model at the plateau (beyond which accuracy of extrapolated estimates is important to capture anticipated projections). Given its good overall fit and on the grounds of parsimony, one knot was considered to be the most suitable choice (Fig. 5a, b). In contrast, estimates with the traditional Weibull model diverged from the external ‘validation’ data, where all patients had progressed by 24 months. By this point in time, traditional methods were already failing to capture the shape of the I-O survival data, leading to an underestimation of the potential gains in survival.
Lastly, extrapolated estimates were checked for realism. Visual inspection of the RCS models over 10 years [4] appeared to show a realistic representation of survival when compared with external long-term data (Fig. 6a–d). At 10 years, extrapolated PFS had declined to around 20%, approximately the level at which the plateau occurred in the OS data pooled from a number of ipilimumab studies [4]. The pooled results have not been superimposed on the 10-year survival projections since they relate to OS rather than PFS.
Extrapolated estimates with traditional and cubic spline methods applied for 10 years to the a combination arm of the ‘training’ data set, b combination arm of the ‘validation’ data set, c ipilimumab arm of the ‘training’ data set, and d ipilimumab arm of the ‘validation’ data set. PFS progression-free survival, RCS restricted cubic spline
4 Discussion
As the assessment of long-term effectiveness and cost-effectiveness is central to decisions made by HTA agencies, it is important that these decisions are informed by appropriate extrapolation of survival, to ensure that calculations of life expectancy and QALYs are reflective of treatment effects. In the appraisal of I-O therapies, the observed pattern of treatment response is not accurately captured by traditional survival functions, which typically exhibit a monotonic trend in hazards. In response, analysts have developed approaches which combine different functional forms or combine observed data with modelled extrapolations. In exploratory modelling, a combination of exponential curves has been found to give a close match to registry data in malignant melanoma [15]. However, the applicability of this for I-O more generally is uncertain as the novelty of therapies may mean that they are not yet represented in registry data.
A variant on the combined functions approach is to append a standard parametric survival function to the complete K–M curve. In one study based on the MDX010-20 trial, a parametric function was used to link K–M data with longer term observational data beyond 5 years [27]. In this case, choice of the point in the tail of the K–M curve at which to begin the extrapolation can become problematic, and it has been argued that applying a survival function to the full range of data is often preferable [23].
An approach which can be relevant in the I-O context given the long-term survival potential and which is worthy of mention here is the cure fraction model, where cure defines the mortality rate of the diseased group relative to the general non-diseased population. Two major model approaches exist, the mixture and non-mixture types, with the former being more widely applied to curable diseases. Such models can offer a useful tool to monitor survival trends and may reduce bias in OS estimates to improve the accuracy of economic value assessments. Cure models have been applied to patient-level data with some success [28] and also to the economic evaluation of ipilimumab in advanced melanoma [29]. However, they require evidence to support an estimate of the fraction of patients ‘cured’ and are therefore potentially more applicable once a biomarker/treatment predictor is established. Methods to date have used matched general population mortality data to determine the fraction of patients who are considered ‘statistically cured’ [28, 29]. For international trials, data limitations regarding availability of suitable background mortality data may cause issues. Furthermore, to accurately identify those who are ‘statistically cured’, mature data are required [29]. As the OS data used in the current investigation were not considered to be sufficiently mature to establish a cure-fraction with confidence, and modelling of PFS is not applicable using cure-rate models, their application in the current study was limited and therefore not undertaken.
The approach to survival modelling based on RCS was found in this study to give a good fit to I-O data drawn from one clinical trial among melanoma patients and performed better than conventional survival functions on the basis of visual inspection and statistical tests. RCS approaches therefore merit consideration alongside other flexible methods as part of the toolkit for the modelling and extrapolation of survival in this therapy area. When using these methods, the analyst needs to address issues related to model selection (Royston [30] suggests strategies for addressing this) and complexity versus data fit. In line with Royston, this study showed that it is possible to find a parsimonious approach with one knot, at little cost in terms of fit, while attempts to optimise knot placement, which is always capable of some improvement, may not achieve a substantial improvement in results [30]. Analytical tractability should not, however, divert attention from questions of clinical plausibility and coherence with the disease process. While spline-based approaches can be considered candidate techniques for survival analysis, they will not always be preferred [20].
4.1 Recommendations
While the most appropriate flexible model and form of the RCS approach will depend on the nature of the data set being analysed, it is recommended that the analyst take a systematic approach to data handling, informed by the steps taken in this paper:
-
1.
Data partitioning: Consistent with existing guidance [17, 19], we recommend randomly partitioning core trial data 1:1 into a ‘training’ and ‘validation’ data set and plotting a 95% CI around the K–M curves to assess uncertainty and heterogeneity between data sets. When the dataset is too limited for division into training and validation data sets (it has been suggested that, for survival time studies, the test sample size should be at least 100 [19]) or data splitting is inconclusive, then bootstrap resampling can be considered as a means of internal validation [17]. Rather than divide the observed data, bootstrapping creates simulated validation data sets by sampling at random with replacement from the observed data. Data splitting was deemed suitable in this example given the sample size, while external validation, which has been proposed as a superior method of validation [19], was also conducted.
-
2.
Data fits: Interpolation with traditional parametric methods applied to the ‘training’ data set is recommended, followed by interpolation using flexible models. In the case of RCS, setting an upper limit on the number of knots (six has been proposed [20], with up to seven tested here) and testing of the sensitivity of results to knot placement are recommended.
-
3.
Goodness of fit: All methods applied to the ‘training’ data set should be assessed by visual inspection of the overall K–M plot and at distinct survival periods with emphasis on the plateau, followed by comparisons of AIC and BIC statistics (these can highlight the need for multiple evaluations before the optimal fit is determined using conventional approaches).
-
4.
Model selection: The preferred model should balance goodness of fit and complexity of the approach while retaining clinical plausibility. Although the lowest AIC and BIC values in this example were associated with the six-knot model (combination therapy) or seven-knot model (monotherapy), the RCS model with one knot gave equivalence of fit for the critical plateau phase of the data. This allowed model complexity and the risk of overfitting to be reduced, as well as improving generalisability (factors to consider alongside goodness of fit and complexity).
-
5.
Hazard plots: Consistent with guidance on the identification of hazard trends [15, 17, 20], we recommend inspection of cumulative and log-cumulative hazards for both treatment arms to support model selection.
-
6.
Consistency: Steps 2–5 should be repeated for the ‘validation’ data set and consistency of results across data sets confirmed.
-
7.
Extrapolation and external validation: It is recommended that data fits be extrapolated to available long-term data (from e.g. registry databases); in this example, validity was assessed on the first 5 years to allow comparison of long-term follow-up [2] followed by long-term extrapolation up to 10 years [4].
4.2 Limitations
The first caveat to note with the analysis is that patient-level data may not always be accessible. In this case, published K–M estimates should be extracted and handled using recognised approaches such as digitally scanning K–M curves [17]. Although RCS models outperformed all model classes in this study, consistency in data fits across the ‘training’ and ‘validation’ data sets may vary. Here, the multiple checkpoints in goodness of fit and validation assessments will aid in justifying the choice of model. Where testing of data fits with AIC or BIC is not possible because these statistics are not applicable, analysts should use other tests or the validations provided in the recommendations above to ensure that the optimal model is selected.
Although our results using combined functions did not produce favourable results, the analysis was limited to the extent that additional parametric functions such as log normal, log-logistic, and Gompertz were not analysed and may have demonstrated different results. Secondly, K–M data were combined with only one parametric function to model survival. Due to the potentially numerous hazard trends displayed, applying additional functions may have resulted in improved fit at the tail, and therefore improved extrapolation estimates. However, on grounds of parsimony, the potential bias arising from small patient numbers [23], and the substantial subjectivity in choosing data points at which to implement separate functions [15], fitting a number of different functions may not be a preferred approach.
When interpreting this analysis, it should be borne in mind that the single outcome measure of PFS has been considered, whereas the development of economic models may involve handling multiple endpoints (e.g. PFS and OS). In this context, flexible (and other) models can potentially provide unrealistic estimates. For example, modelled PFS can exceed modelled OS under extrapolation, thus highlighting the importance of clinical plausibility. Contributing factors to the crossing of OS and PFS curves may be immature OS data or inclusion of low patient numbers in the last few observations. However, if a systematic approach is taken to both endpoints simultaneously, this can be handled by selecting different models for different endpoints, removing the last few observations [20] or attaching greater weight to the data with the longest follow-up. Additionally, sensitivity analysis with data fits for different treatment durations (i.e. trial period vs follow-up data) can be conducted.
5 Conclusions
Survival analyses presented in economic evaluations of I-O continue to be performed using traditional parametric methods, which do not take into account the mode of action of I-O therapies [31, 32], thus failing to capture the ‘tail’ of the survival curve and treatment pathway in extrapolated estimates. This implies that the clinical value of such compounds may be underestimated, giving rise to misleading estimates of cost-effectiveness. This study has shown that spline-based models using a limited number of knots can provide an acceptable fit to trial data and generate extrapolated estimates supported by longer term evidence, with results that are stable in response to changes in knot placement. By following a robust methodology and validating findings when using more flexible models, of which spline-based methods are an example, subjectivity and uncertainty surrounding the assumptions required for more complex approaches can be minimised. These models can be considered a useful addition to the analytical tools available to estimate survival in I-O. The applicability of these findings to other conditions and other treatment regimens (e.g. chemo + I-O combinations) requires further exploration.
Data Availability Statement
The analysis reported in this study uses patient-level data from the CheckMate 067 trial. The patient-level data are not publicly available, but the results of the trial have been reported in a number of publications. The trial results supporting the findings of this analysis are presented graphically within the article. The survival analysis was implemented in Stata, with the RCS models fitted using the stpm2 command.
Notes
Traditionally, significant tumour growth implies disease progression and treatment failure. In I-O, an apparent increase in tumour size or development of new growths revealed by scans of the tumour site are sometimes found to be an infiltration of the host’s tumour cells (pseudo-progression) rather than disease progression. These infiltrates can subsequently clear, with a favourable clinical response being reported.
References
Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–26.
Hodi FS, Day SJO, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Ph D, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23.
Food and Drug Administration (United States). FY 2011 innovative drug approvals. Silver Spring: FDA; 2011.
Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, Patt D, Chen T-T, Berman DM, Wolchok JD. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33:1889–94.
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. doi:10.1016/j.ejca.2008.10.026.
Ades F, Yamaguchi N. WHO, RECIST, and immune-related response criteria: is it time to revisit pembrolizumab results? Ecancermedicalscience. 2015;9:9–12.
Wolchok JD, Hoos A, O’Day S, Weber JS, Hamid O, Lebbé C, Maio M, Binder M, Bohnsack O, Nichol G, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–20.
Hodi FS, Hwu WJ, Kefford R, Weber JS, Daud A, Hamid O, Patnaik A, Ribas A, Robert C, Gangadhar TC, et al. Evaluation of immune-related response criteria and RECIST v1.1 in patients with advanced melanoma treated with pembrolizumab. J Clin Oncol. 2016;34:1510–7.
Karydis I, Chan PY, Wheater M, Arriola E, Szlosarek PW, Ottensmeier CH. Clinical activity and safety of pembrolizumab in ipilimumab pre-treated patients with uveal melanoma. Oncoimmunology. 2016;5:e1143997. doi:10.1080/2162402X.2016.1143997.
Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34.
Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–32. doi:10.1056/NEJMoa1503093.
McDermott DF, Drake CG, Sznol M, Choueiri TK, Powderly JD, Smith DC, Brahmer JR, Carvajal RD, Hammers HJ, Puzanov I, et al. Survival, durable response, and long-term safety in patients with previously treated advanced renal cell carcinoma receiving nivolumab. J Clin Oncol. 2015;33:2013–20.
Chen T-T. Statistical issues and challenges in immuno-oncology. J Immunother Cancer. 2013;1:18.
Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–30.
Bagust A, Beale S. Survival analysis and extrapolation modeling of time-to-event clinical trial data for economic evaluation: an alternative approach. Med Decis Mak. 2014;34:343–51.
Giannopoulou C, Siderios E, Wade R, Moe-Byrne T, Eastwood A, McKenna C. Ipilimumab for previously untreated unresectable malignant melanoma: a critique of the evidence. PharmacoEconomics. 2015;33:1269–79.
Latimer N. NICE DSU Technical Support Document 14: survival analysis for economic evaluations alongside clinical trials-extrapolation with patient-level data. Sheffield: University of Sheffield; 2013.
NICE. Committee papers Single Technology Appraisal STA nivolumab for treating advanced (unresectable or metastatic) melanoma [ID848]. London: 2016.
Harrell FE. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. 2nd ed. Cham: Springer; 2015.
Tremblay G, Haines P, Briggs A. A criterion-based approach for the systematic and transparent extrapolation of clinical trial survival data. JHEOR. 2015;2:147–60.
Jackson C, Stevens J, Ren S, Latimer N, Bojke L, Manca A. Extrapolating survival from randomized trials using external data: a review of methods. Med Decis Mak. 2017;37(4):377–90.
NICE. Committee papers Single Technology Appraisal nivolumab for treating advanced (unresectable or metastatic) melanoma [ID845]. London: National Institute for Health and Care Excellence; 2016.
Davies A, Briggs A, Schneider J, Levy A, Ebeid O, Wagner S, Kotapati S, Ramsey S. The ends justify the mean: outcome measures for estimating the value of new cancer therapies. Health Outcomes Res Med. 2012;3:e25–36. doi:10.1016/j.ehrm.2012.01.001.
Royston P, Lambert PC. Flexible parametric survival analysis using Stata: beyond the Cox model. College Station: Stata Press, USA; 2011.
Rutherford MJ, Crowther MJ, Lambert PC. Using restricted cubic splines to approximate complex hazard functions in the analysis of time-to-event data: a simulation study. J Stat Comput Simul. 2015;85:777–93.
Lambert P, Royston P. Further development of flexible parametric models for survival analysis. Stata J. 2009;9:265–90.
Larkin J, Hatswell AJ, Nathan P, Lebmeier M, Lee D. The predicted impact of ipilimumab usage on survival in previously treated advanced or metastatic melanoma in the UK. PLoS One. 2015;10:1–11.
Lambert PC, Thompson JR, Weston CL, Dickman PW. Estimating and modeling the cure fraction in population-based cancer survival analysis. Biostatistics. 2007;8:576–94.
Othus M, Bansal A, Koepl L, Wagner S, Ramsey S. Accounting for cured patients in cost-effectiveness analysis. Value Health. 2017;20:705–9.
Royston P. Flexible parametric alternatives to the Cox model, and more. Stata J [Internet] 2001; 1:1–28. http://ideas.repec.org/a/tsj/stataj/v7y2007i4p465-506.html.
Jensen IS, Zacherle E, Blanchette CM, Zhang J, Yin W. Evaluating cost benefits of combination therapies for advanced melanoma. Drugs Context. 2016;5:1–14.
Wang J, Chmielowski B, Pellissier J, Xu R, Stevinson K, Liu FX. Cost-effectiveness of pembrolizumab versus ipilimumab in ipilimumab-naive patients with advanced melanoma in the United States. J Manag Care Spec Pharm. 2017;23:184–94.
Acknowledgements
The authors are grateful to Professor Mondher Toumi and three anonymous referees for valuable comments on an earlier draft of this paper, and to Clive Pritchard of Wickenstones for writing and editorial assistance.
Author information
Authors and Affiliations
Contributions
All authors contributed to study conception and design and drafted the manuscript. Ian Koblbauer and Najida Begum performed the survival analysis. Eddie Gibson is the guarantor of the research.
Corresponding author
Ethics declarations
Funding
This research was funded by Bristol-Myers Squibb.
Conflict of interest
Ariadna Juarez-Garcia, Michael Lees, Amir Abbas Tahami Monfared, David Tyas and Yong Yuan were employed by Bristol-Myers Squibb (BMS). Najida Begum, Eddie Gibson, and Ian Koblbauer were employed by Wickenstones Ltd, who were funded by BMS to undertake the research. George Dranitsaris, Danny Liew and Phil McEwan have received consultancy fees and have been reimbursed for travel expenses to attend advisory board meetings related to this research.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gibson, E., Koblbauer, I., Begum, N. et al. Modelling the Survival Outcomes of Immuno-Oncology Drugs in Economic Evaluations: A Systematic Approach to Data Analysis and Extrapolation. PharmacoEconomics 35, 1257–1270 (2017). https://doi.org/10.1007/s40273-017-0558-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40273-017-0558-5