Abstract
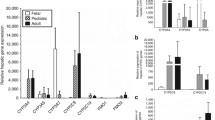
The aim of this review is to discuss our current understanding of the developmental changes of the drug-metabolizing enzyme cytochrome P450 (CYP) 3A and its impact on drug therapy. In the last 10 years, several methods have been used to study the ontogeny of specific CYP3A isoforms in vitro and in vivo. Although most studies confirm previous findings that CYP3A4/5 activity is low at birth and reaches adult values in the first years of life, there are still important gaps in our knowledge of the exact developmental patterns of individual CYP3A isoforms, especially in this age range. Moreover, most in vivo clinical studies have also failed to cover the whole pediatric age range. To date, this information gap still hampers the design of age-specific dosing guidelines of CYP3A substrate drugs, especially in neonates and infants. Innovative study methods, including opportunistic sampling and sensitive analytical assays used in combination with physiologically based pharmacokinetics, and population pharmacokinetic model concepts may help to improve our understanding of the ontogeny of CYP3A and aid the application of this knowledge in clinical practice.


Similar content being viewed by others
References
Finta C, Zaphiropoulos PG. The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons. Gene. 2000;260(1–2):13–23.
Domanski TL, Finta C, Halpert JR, et al. cDNA cloning and initial characterization of CYP3A43, a novel human cytochrome P450. Mol Pharmacol. 2001;59(2):386–92.
Watkins PB. Noninvasive tests of CYP3A enzymes. Pharmacogenetics. 1994;4(4):171–84.
Gellner K, Eiselt R, Hustert E, et al. Genomic organization of the human CYP3A locus: identification of a new, inducible CYP3A gene. Pharmacogenetics. 2001;11(2):111–21.
Lamba JK, Lin YS, Schuetz EG, et al. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54(10):1271–94.
Sim SC, Edwards RJ, Boobis AR, et al. CYP3A7 protein expression is high in a fraction of adult human livers and partially associated with the CYP3A7*1C allele. Pharmacogenet Genomics. 2005;15(9):625–31.
Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17.
Ratanasavanh D, Beaune P, Morel F, et al. Intralobular distribution and quantitation of cytochrome P-450 enzymes in human liver as a function of age. Hepatology. 1991;13(6):1142–51.
Kolars JC, Lown KS, Schmiedlin-Ren P, et al. CYP3A gene expression in human gut epithelium. Pharmacogenetics. 1994;4(5):247–59.
Watkins PB, Wrighton SA, Schuetz EG, et al. Identification of glucocorticoid-inducible cytochromes P-450 in the intestinal mucosa of rats and man. J Clin Invest. 1987;80(4):1029–36.
Westlind A, Malmebo S, Johansson I, et al. Cloning and tissue distribution of a novel human cytochrome p450 of the CYP3A subfamily, CYP3A43. Biochem Biophys Res Commun. 2001;281(5):1349–55.
Vyhlidal CA, Gaedigk R, Leeder JS. Nuclear receptor expression in fetal and pediatric liver: correlation with CYP3A expression. Drug Metab Dispos. 2006;34(1):131–7.
Burk O, Tegude H, Koch I, et al. Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine. J Biol Chem. 2002;277(27):24280–8.
Paine MF, Khalighi M, Fisher JM, et al. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther. 1997;283(3):1552–62.
Shimada T, Yamazaki H, Mimura M, et al. Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs. Drug Metab Dispos. 1996;24(5):515–22.
Hall SD, Thummel KE, Watkins PB, et al. Molecular and physical mechanisms of first-pass extraction. Drug Metab Dispos. 1999;27(2):161–6.
Ozdemir M, Crewe KH, Tucker GT, et al. Assessment of in vivo CYP2D6 activity: differential sensitivity of commonly used probes to urine pH. J Clin Pharmacol. 2004;44(12):1398–404.
Elens L, Becker ML, Haufroid V, et al. Novel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin-mediated cholesterol reduction in The Rotterdam Study. Pharmacogenet Genomics. 2011;21(12):861–6.
Elens L, van Schaik RH, Panin N, et al. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics. 2011;12(10):1383–96.
Lee SJ, Goldstein JA. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics. 2005;6(4):357–71.
Perera MA. The missing linkage: what pharmacogenetic associations are left to find in CYP3A? Expert Opin Drug Metab Toxicol. 2010;6(1):17–28.
Schroder A, Klein K, Winter S, et al. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 2013:13(1):12–20.
Yang X, Zhang B, Molony C, et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20(8):1020–36.
Klein K, Thomas M, Winter S, et al. PPARA: a novel genetic determinant of CYP3A4 in vitro and in vivo. Clin Pharmacol Ther. 2012;91(6):1044–52.
Gibbs MA, Thummel KE, Shen DD, et al. Inhibition of cytochrome P-450 3A (CYP3A) in human intestinal and liver microsomes: comparison of K i values and impact of CYP3A5 expression. Drug Metab Dispos. 1999;27(2):180–7.
Haehner BD, Gorski JC, Vandenbranden M, et al. Bimodal distribution of renal cytochrome P450 3A activity in humans. Mol Pharmacol. 1996;50(1):52–9.
Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–91.
Maezawa K, Matsunaga T, Takezawa T, et al. Cytochrome P450 3As gene expression and testosterone 6beta-hydroxylase activity in human fetal membranes and placenta at full term. Biol Pharm Bull. 2010;33(2):249–54.
van Schaik RH, van der Heiden IP, van den Anker JN, et al. CYP3A5 variant allele frequencies in Dutch Caucasians. Clin Chem. 2002;48(10):1668–71.
Macphee IA, Fredericks S, Mohamed M, et al. Tacrolimus pharmacogenetics: the CYP3A5*1 allele predicts low dose-normalized tacrolimus blood concentrations in whites and South Asians. Transplantation. 2005;79(4):499–502.
Zhao Y, Song M, Guan D, et al. Genetic polymorphisms of CYP3A5 genes and concentration of the cyclosporine and tacrolimus. Transpl Proc. 2005;37(1):178–81.
Frohlich M, Hoffmann MM, Burhenne J, et al. Association of the CYP3A5 A6986G (CYP3A5*3) polymorphism with saquinavir pharmacokinetics. Br J Clin Pharmacol. 2004;58(4):443–4.
Mouly SJ, Matheny C, Paine MF, et al. Variation in oral clearance of saquinavir is predicted by CYP3A5*1 genotype but not by enterocyte content of cytochrome P450 3A5. Clin Pharmacol Ther. 2005;78(6):605–18.
Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15(6):415–21.
Kivisto KT, Niemi M, Schaeffeler E, et al. Lipid-lowering response to statins is affected by CYP3A5 polymorphism. Pharmacogenetics. 2004;14(8):523–5.
Inoue K, Inazawa J, Nakagawa H, et al. Assignment of the human cytochrome P-450 nifedipine oxidase gene (CYP3A4) to chromosome 7 at band q22.1 by fluorescence in situ hybridization. Jpn J Hum Genet. 1992;37(2):133–8.
Komori M, Nishio K, Ohi H, et al. Molecular cloning and sequence analysis of cDNA containing the entire coding region for human fetal liver cytochrome P-450. J Biochem. 1989;105(2):161–3.
Lacroix D, Sonnier M, Moncion A, et al. Expression of CYP3A in the human liver—evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem. 1997;247(2):625–34.
Nishimura M, Yaguti H, Yoshitsugu H, et al. Tissue distribution of mRNA expression of human cytochrome P450 isoforms assessed by high-sensitivity real-time reverse transcription PCR. Yakugaku Zasshi. 2003;123(5):369–75.
Stevens JC, Hines RN, Gu C, et al. Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther. 2003;307(2):573–82.
Kitada M, Kamataki T, Itahashi K, et al. P-450 HFLa, a form of cytochrome P-450 purified from human fetal livers, is the 16 alpha-hydroxylase of dehydroepiandrosterone 3-sulfate. J Biol Chem. 1987;262(28):13534–7.
Leeder JS, Gaedigk R, Marcucci KA, et al. Variability of CYP3A7 expression in human fetal liver. J Pharmacol Exp Ther. 2005;314(2):626–35.
Rodriguez-Antona C, Jande M, Rane A, et al. Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers. Clin Pharmacol Ther. 2005;77(4):259–70.
Daly AK. Significance of the minor cytochrome P450 3A isoforms. Clin Pharmacokinet. 2006;45(1):13–31.
Ohmori S, Nakasa H, Asanome K, et al. Differential catalytic properties in metabolism of endogenous and exogenous substrates among CYP3A enzymes expressed in COS-7 cells. Biochim Biophys Acta. 1998;1380(3):297–304.
de Wildt SN, Ito S, Koren G. Challenges for drug studies in children: CYP3A phenotyping as example. Drug Discov Today. 2009;14(1–2):6–15.
Hakkola J, Tanaka E, Pelkonen O. Developmental expression of cytochrome P450 enzymes in human liver. Pharmacol Toxicol. 1998;82(5):209–17.
Rich KJ, Boobis AR. Expression and inducibility of P450 enzymes during liver ontogeny. Microsc Res Tech. 1997;39(5):424–35.
Stevens JC. New perspectives on the impact of cytochrome P450 3A expression for pediatric pharmacology. Drug Discov Today. 2006;11(9–10):440–5.
Schuetz JD, Beach DL, Guzelian PS. Selective expression of cytochrome P450 CYP3A mRNAs in embryonic and adult human liver. Pharmacogenetics. 1994;4(1):11–20.
Greuet J, Pichard L, Bonfils C, et al. The fetal specific gene CYP3A7 is inducible by rifampicin in adult human hepatocytes in primary culture. Biochem Biophys Res Commun. 1996;225(2):689–94.
Hakkola J, Pasanen M, Purkunen R, et al. Expression of xenobiotic-metabolizing cytochrome P450 forms in human adult and fetal liver. Biochem Pharmacol. 1994;48(1):59–64.
Pearce RE, McIntyre CJ, Madan A, et al. Effects of freezing, thawing, and storing human liver microsomes on cytochrome P450 activity. Arch Biochem Biophys. 1996;331(2):145–69.
Hakkola J, Raunio H, Purkunen R, et al. Cytochrome P450 3A expression in the human fetal liver: evidence that CYP3A5 is expressed in only a limited number of fetal livers. Biol Neonate. 2001;80(3):193–201.
Wrighton SA, Brian WR, Sari MA, et al. Studies on the expression and metabolic capabilities of human liver cytochrome P450IIIA5 (HLp3). Mol Pharmacol. 1990;38(2):207–13.
Gorski JC, Hall SD, Jones DR, et al. Regioselective biotransformation of midazolam by members of the human cytochrome P450 3A (CYP3A) subfamily. Biochem Pharmacol. 1994;47(9):1643–53.
Walsky RL, Obach RS, Hyland R, et al. Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug Metab Dispos. 2012;40(9):1686–97.
Yang HY, Lee QP, Rettie AE, et al. Functional cytochrome P4503A isoforms in human embryonic tissues: expression during organogenesis. Mol Pharmacol. 1994;46(5):922–8.
Aleksa K, Matsell D, Krausz K, et al. Cytochrome P450 3A and 2B6 in the developing kidney: implications for ifosfamide nephrotoxicity. Pediatr Nephrol. 2005;20(7):872–85.
Fakhoury M, de Beaumais T, Guimiot F, et al. mRNA expression of MDR1 and major metabolising enzymes in human fetal tissues. Drug Metab Pharmacokinet. 2009;24(6):529–36.
Fakhoury M, Litalien C, Medard Y, et al. Localization and mRNA expression of CYP3A and P-glycoprotein in human duodenum as a function of age. Drug Metab Dispos. 2005;33(11):1603–7.
Johnson TN, Tanner MS, Taylor CJ, et al. Enterocytic CYP3A4 in a paediatric population: developmental changes and the effect of coeliac disease and cystic fibrosis. Brit J Clin Pharmacol. 2001;51(5):451–60.
Treluyer JM, Jacqz-Aigrain E, Alvarez F, et al. Expression of CYP2D6 in developing human liver. Eur J Biochem. 1991;202(2):583–8.
Fukudo M, Yano I, Masuda S, et al. Population pharmacokinetic and pharmacogenomic analysis of tacrolimus in pediatric living-donor liver transplant recipients. Clin Pharmacol Ther. 2006;80(4):331–45.
Fukudo M, Yano I, Yoshimura A, et al. Impact of MDR1 and CYP3A5 on the oral clearance of tacrolimus and tacrolimus-related renal dysfunction in adult living-donor liver transplant patients. Pharmacogenet Genomics. 2008;18(5):413–23.
Schuetz JD, Kauma S, Guzelian PS. Identification of the fetal liver cytochrome CYP3A7 in human endometrium and placenta. J Clin Invest. 1993;92(2):1018–24.
Kirwan C, MacPhee I, Philips B. Using drug probes to monitor hepatic drug metabolism in critically ill patients: midazolam, a flawed but useful tool for clinical investigation of CYP3A activity? Expert Opin Drug Metab Toxicol. 2010;6(6):761–71.
Kashuba AD, Nafziger AN, Kearns GL, et al. Limitations of dextromethorphan N-demethylation as a measure of CYP3A activity. Pharmacogenetics. 1999;9(4):453–62.
Lowry JA, Kearns GL, Abdel-Rahman SM, et al. Cisapride: a potential model substrate to assess cytochrome P4503A4 activity in vivo. Clin Pharmacol Ther. 2003;73(3):209–22.
de Wildt SN, Kearns GL, Hop WC, et al. Pharmacokinetics and metabolism of intravenous midazolam in preterm infants. Clin Pharmacol Ther. 2001;70(6):525–31.
de Wildt SN, de Hoog M, Vinks AA, et al. Population pharmacokinetics and metabolism of midazolam in pediatric intensive care patients. Crit Care Med. 2003;31(7):1952–8.
Allegaert K, Van den Anker JN, Debeer A, et al. Maturational changes in the in vivo activity of CYP3A4 in the first months of life. Int J Clin Pharmacol Ther. 2006;44(7):303–8.
Blake MJ, Abdel-Rahman SM, Pearce RE, et al. Effect of diet on the development of drug metabolism by cytochrome P-450 enzymes in healthy infants. Pediatr Res. 2006;60(6):717–23.
de Wildt SN, Berns MJ, van den Anker JN. 13C-erythromycin breath test as a noninvasive measure of CYP3A activity in newborn infants: a pilot study. Ther Drug Monit. 2007;29(2):225–30.
de Wildt SN, Kearns GL, Hop WC, et al. Pharmacokinetics and metabolism of oral midazolam in preterm infants. Br J Clin Pharmacol. 2002;53(4):390–2.
Shimada T, Yamazaki H, Mimura M, et al. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270(1):414–23.
Thummel KE, Shen DD, Podoll TD, et al. Use of midazolam as a human cytochrome P450 3A probe: I. In vitro–in vivo correlations in liver transplant patients. J Pharmacol Exp Ther. 1994;271(1):549–56.
Wang RW, Newton DJ, Liu NY, et al. Inhibitory anti-CYP3A4 peptide antibody: mapping of inhibitory epitope and specificity toward other CYP3A isoforms. Drug Metab Dispos. 1999;27(2):167–72.
Burtin P, Jacqz-Aigrain E, Girard P, et al. Population pharmacokinetics of midazolam in neonates. Clin Pharmacol Ther. 1994;56(6 Pt 1):615–25.
Jacqz-Aigrain E, Wood C, Robieux I. Pharmacokinetics of midazolam in critically ill neonates. Eur J Clin Pharmacol. 1990;39(2):191–2.
Jacqz-Aigrain E, Daoud P, Burtin P, et al. Pharmacokinetics of midazolam during continuous infusion in critically ill neonates. Eur J Clin Pharmacol. 1992;42(3):329–32.
Williams JA, Ring BJ, Cantrell VE, et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30(8):883–91.
Rey E, Delaunay L, Pons G, et al. Pharmacokinetics of midazolam in children: comparative study of intranasal and intravenous administration. Eur J Clin Pharmacol. 1991;41(4):355–7.
Reed MD, Rodarte A, Blumer JL, et al. The single-dose pharmacokinetics of midazolam and its primary metabolite in pediatric patients after oral and intravenous administration. J Clin Pharmacol. 2001;41(12):1359–69.
Jones RD, Chan K, Roulson CJ, et al. Pharmacokinetics of flumazenil and midazolam. Br J Anaesth. 1993;70(3):286–92.
Tolia V, Brennan S, Aravind MK, et al. Pharmacokinetic and pharmacodynamic study of midazolam in children during esophagogastroduodenoscopy. J Pediatr. 1991;119(3):467–71.
Hartwig S, Roth B, Theisohn M. Clinical experience with continuous intravenous sedation using midazolam and fentanyl in the paediatric intensive care unit. Eur J Pediatr. 1991;150(11):784–8.
Peeters MY, Prins SA, Knibbe CA, et al. Propofol pharmacokinetics and pharmacodynamics for depth of sedation in nonventilated infants after major craniofacial surgery. Anesthesiology. 2006;104(3):466–74.
Peeters MY, Prins SA, Knibbe CA, et al. Pharmacokinetics and pharmacodynamics of midazolam and metabolites in nonventilated infants after craniofacial surgery. Anesthesiology. 2006;105(6):1135–46.
Greenblatt DJ, Ehrenberg BL, Gunderman J, et al. Pharmacokinetic and electroencephalographic study of intravenous diazepam, midazolam, and placebo. Clin Pharmacol Ther. 1989;45(4):356–65.
Mandema JW, Tuk B, van Steveninck AL, et al. Pharmacokinetic-pharmacodynamic modeling of the central nervous system effects of midazolam and its main metabolite alpha-hydroxymidazolam in healthy volunteers. Clin Pharmacol Ther. 1992;51(6):715–28.
Anderson BJ, Larsson P. A maturation model for midazolam clearance. Paediatr Anaesth. 2011;21(3):302–8.
Vet NJ, de Hoog M, Tibboel D, et al. The effect of critical illness and inflammation on midazolam therapy in children. Pediatr Crit Care Med. 2012;13(1):e48–50.
Hughes J, Gill AM, Mulhearn H, et al. Steady-state plasma concentrations of midazolam in critically ill infants and children. Ann Pharmacother. 1996;30(1):27–30.
Mathews HM, Carson IW, Lyons SM, et al. A pharmacokinetic study of midazolam in paediatric patients undergoing cardiac surgery. Br J Anaesth. 1988;61(3):302–7.
Fromm MF, Busse D, Kroemer HK, et al. Differential induction of prehepatic and hepatic metabolism of verapamil by rifampin. Hepatology. 1996;24(4):796–801.
Hebert MF, Roberts JP, Prueksaritanont T, et al. Bioavailability of cyclosporine with concomitant rifampin administration is markedly less than predicted by hepatic enzyme induction. Clin Pharmacol Ther. 1992;52(5):453–7.
Holtbecker N, Fromm MF, Kroemer HK, et al. The nifedipine–rifampin interaction. Evidence for induction of gut wall metabolism. Drug Metab Dispos. 1996;24(10):1121–3.
Thummel KE, O’Shea D, Paine MF, et al. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clin Pharmacol Ther. 1996;59(5):491–502.
Wu CY, Benet LZ, Hebert MF, et al. Differentiation of absorption and first-pass gut and hepatic metabolism in humans: studies with cyclosporine. Clin Pharmacol Ther. 1995;58(5):492–7.
Lown KS, Ghosh M, Watkins PB. Sequences of intestinal and hepatic cytochrome P450 3A4 cDNAs are identical. Drug Metab Dispos. 1998;26(2):185–7.
Smith MT, Eadie MJ, Brophy TO. The pharmacokinetics of midazolam in man. Eur J Clin Pharmacol. 1981;19(4):271–8.
Marshall J, Rodarte A, Blumer J, et al. Pediatric pharmacodynamics of midazolam oral syrup. Pediatric Pharmacology Research Unit Network. J Clin Pharmacol. 2000;40(6):578–89.
Maclennan S, Augood C, Cash-Gibson L, et al. Cisapride treatment for gastro-oesophageal reflux in children. Cochrane Database Syst Rev. 2010(4):CD002300.
Preechagoon Y, Charles B, Piotrovskij V, et al. Population pharmacokinetics of enterally administered cisapride in young infants with gastro-oesophageal reflux disease. Brit J Clin Pharmacol. 1999;48(5):688–93.
Kearns GL, Robinson PK, Wilson JT, et al. Cisapride disposition in neonates and infants: in vivo reflection of cytochrome P450 3A4 ontogeny. Clin Pharmacol Ther. 2003;74(4):312–25.
Blake MJ, Gaedigk A, Pearce RE, et al. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin Pharmacol Ther. 2007;81(4):510–6.
Johnson TN, Tucker GT, Rostami-Hodjegan A. Development of CYP2D6 and CYP3A4 in the first year of life. Clin Pharmacol Ther. 2008;83(5):670–1.
Rostami-Hodjegan A, Kroemer HK, Tucker GT. In-vivo indices of enzyme activity: the effect of renal impairment on the assessment of CYP2D6 activity. Pharmacogenetics. 1999;9(3):277–86.
Johnson TN, Thomson M. Intestinal metabolism and transport of drugs in children: the effects of age and disease. J Pediatric Gastroenterol Nutr. 2008;47(1):3–10.
Ferraresso M, Tirelli A, Ghio L, et al. Influence of the CYP3A5 genotype on tacrolimus pharmacokinetics and pharmacodynamics in young kidney transplant recipients. Pediatr Transpl. 2007;11(3):296–300.
de Wildt SN, van Schaik RH, Soldin OP, et al. The interactions of age, genetics, and disease severity on tacrolimus dosing requirements after pediatric kidney and liver transplantation. Eur J Clin Pharmacol. 2011.
Gijsen V, Mital S, van Schaik RH, et al. Age and CYP3A5 genotype affect tacrolimus dosing requirements after transplant in pediatric heart recipients. J Heart Lung Transpl. 2011;30(12):1352–9.
Zhao W, Elie V, Roussey G, et al. Population pharmacokinetics and pharmacogenetics of tacrolimus in de novo pediatric kidney transplant recipients. Clin Pharmacol Ther. 2009;86(6):609–18.
Terrazzino S, Quaglia M, Stratta P, et al. The effect of CYP3A5 6986A>G and ABCB1 3435C>T on tacrolimus dose-adjusted trough levels and acute rejection rates in renal transplant patients: a systematic review and meta-analysis. Pharmacogenet Genomics. 2012;22(8):642–5.
Drocourt L, Ourlin JC, Pascussi JM, et al. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem. 2002;277(28):25125–32.
Gibson GG, Plant NJ, Swales KE, et al. Receptor-dependent transcriptional activation of cytochrome P4503A genes: induction mechanisms, species differences and interindividual variation in man. Xenobiotica. 2002;32(3):165–206.
Goodwin B, Redinbo MR, Kliewer SA. Regulation of cyp3a gene transcription by the pregnane x receptor. Annu Rev Pharmacol Toxicol. 2002;42:1–23.
Pascussi JM, Gerbal-Chaloin S, Drocourt L, et al. The expression of CYP2B6, CYP2C9 and CYP3A4 genes: a tangle of networks of nuclear and steroid receptors. Biochim Biophys Acta. 2003;1619(3):243–53.
Chartier FL, Bossu JP, Laudet V, et al. Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene. 1994;147(2):269–72.
Tirona RG, Lee W, Leake BF, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med. 2003;9(2):220–4.
Gerbal-Chaloin S, Daujat M, Pascussi JM, et al. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem. 2002;277(1):209–17.
Kacevska M, Ivanov M, Wyss A, et al. DNA methylation dynamics in the hepatic CYP3A4 gene promoter. Biochimie. 2012;94(11):2338–44.
Goodwin B, Hodgson E, D’Costa DJ, et al. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol. 2002;62(2):359–65.
Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol. 1999;56(6):1329–39.
Hines RN. Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol. 2007;21(4):169–75.
Payne K, Mattheyse FJ, Liebenberg D, et al. The pharmacokinetics of midazolam in paediatric patients. Eur J Clin Pharmacol. 1989;37(3):267–72.
Ince I, de Wildt SN, Peeters MY, et al. Critical illness is a major determinant of midazolam clearance in children aged 1 month to 17 years. Ther Drug Monit. 2012;34(4):381–9.
Vet NJ, de Hoog M, Tibboel D, et al. The effect of inflammation on drug metabolism: a focus on pediatrics. Drug Discov Today. 2011;16(9–10):435–42.
Carcillo JA, Doughty L, Kofos D, et al. Cytochrome P450 mediated-drug metabolism is reduced in children with sepsis-induced multiple organ failure. Intensive Care Med. 2003;29(6):980–4.
Kearns GL, Abdel-Rahman SM, Alander SW, et al. Developmental pharmacology—drug disposition, action, and therapy in infants and children. N Engl J Med. 2003;349(12):1157–67.
Björkman S. Prediction of cytochrome p450-mediated hepatic drug clearance in neonates, infants and children: how accurate are available scaling methods? Clin Pharmacokinet. 2006;45(1):1–11.
Edginton AN, Schmitt W, Voith B, et al. A mechanistic approach for the scaling of clearance in children. Clin Pharmacokinet. 2006;45(7):683–704.
Johnson TN, Rostami-Hodjegan A, Tucker GT. Prediction of the clearance of eleven drugs and associated variability in neonates, infants and children. Clin Pharmacokinet. 2006;45(9):931–56.
Ince I, de Wildt SN, Tibboel D, et al. Tailor-made drug treatment for children: creation of an infrastructure for data-sharing and population PK-PD modeling. Drug Discov Today. 2009;14(5–6):316–20.
Krekels EHJ, Johnson TN, den Hoedt SM, et al. Top–down modeling meets bottom–up modeling: the physiological and physicochemical basis for the ontogeny of UGT2B7-mediated drug glucuronidation. 2012. http://www.page-meeting.org/?abstract=2369.
Leong R, Vieira ML, Zhao P, et al. Regulatory experience with physiologically based pharmacokinetic modeling for pediatric drug trials. Clin Pharmacol Ther. 2012;91(5):926–31.
Schaefer O, Ohtsuki S, Kawakami H, et al. Absolute quantification and differential expression of drug transporters, cytochrome P450 enzymes, and UDP-glucuronosyltransferases in cultured primary human hepatocytes. Drug Metab Dispos. 2012;40(1):93–103.
Muchohi SN, Kokwaro GO, Ogutu BR, et al. Pharmacokinetics and clinical efficacy of midazolam in children with severe malaria and convulsions. Br J Clin Pharmacol. 2008;66(4):529–38.
Acknowledgments
This work was performed within the framework of Dutch Top Institute Pharma project number D2-104. Dr. de Wildt’s research is supported by Erasmus MC and ZonMW clinical fellowships. The work of C.A.J. Knibbe is supported by the Innovational Research Incentives Scheme (Veni grant, July 2006) of the Dutch Organization for Scientific Research (NWO). Meindert Danhof and Ibrahim Ince have declared no conflicts of interest that are directly relevant to the content of this review.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ince, I., Knibbe, C.A.J., Danhof, M. et al. Developmental Changes in the Expression and Function of Cytochrome P450 3A Isoforms: Evidence from In Vitro and In Vivo Investigations. Clin Pharmacokinet 52, 333–345 (2013). https://doi.org/10.1007/s40262-013-0041-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-013-0041-1