Abstract
Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), Middle East Respiratory Syndrome coronavirus (MERS-CoV) and the novel SARS-CoV-2 evade the host innate immunity, and subsequently the adaptive immune response, employing one protease called Papain-like protease (PLpro). The PLpro and the 3CL main protease are responsible for the cleavage of the polyproteins encoded by the + sense RNA genome of the virus to produce several non-structured proteins (NSPs). However, the PLpro also performs deubiquitination and deISGylation of host proteins and signaling molecules, and thus antagonize the host innate immune response, since ubiquitination and ISGylation are critical processes which invoke host’s antiviral immune responses. Thus, to maintain host antiviral defense, inhibition of the PLpro is the primary therapeutic strategy. Furthermore, inhibition of the enzyme prevents replication of the virus. The present study employs molecular modeling approaches to determine potential of different approved and repurposed drugs and other compounds as inhibitors of the SARS-CoV-2 PLpro. The results of the study demonstrated that drugs like Stallimycin, and known protease inhibitors including Telaprevir, Grazoprevir and Boceprevir, were highly potent in inhibiting the enzyme. In addition, several plant-derived polyphenols, including Corylifol A and Kazinol J, were found to be potent inhibitors. Based on the findings, we suggest that clinical trials be initiated with these inhibitors. So far, PLpro inhibition has been given less attention as a strategy to contain COVID-19 pandemic, and thus the present study is of high significance and has therapeutic implications in containing the pandemic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has already taken a death toll of 10,87,069 people and infected over 38,202,956 across the globe, as of October 15, 2020 (https://covid19.who.int/). The coronavirus (CoV), named Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), affects the respiratory tract leading to cough, fever, shortness of breath and pneumonia, and the patients often need life support systems (Chen et al. 2020). Ever since the first case was reported in December (2019) from Wuhan (Hubei province, China) (Lu et al. 2020a), researchers across the globe are striving to develop a therapeutic intervention to control the pandemic. However, instead of the time consuming de novo drug discovery, repurposing of the approved drugs has been investigated more (Guo 2020). This has been facilitated by elucidation of the viral genome sequence and enzyme structures, which are the basis for drug discovery research (Baker 2020; Cunningham et al. 2020). Further, knowledge gathered from studies on other related viruses including SARS-CoV, MERS-CoV and Hepatitis C virus (HCV), has been of great use. Molecular modeling (including molecular docking) tools have been the most useful tools in identifying potential drugs against COVID-19, and so far more focus has been given on main protease (3CLpro) and RNA-dependent RNA polymerase (RdRp) of the virus.
The SARS-CoV-2 has a positive sense single stranded RNA genome of 29.9 kb (Lu et al. 2020b), comprising of 6–11 open reading frames (ORFs) (Guo 2020). The first ORF encodes 16 nonstructural proteins (NSPs) including two cysteine proteases, viz. papain-like protease (NSP3), chymotrypsin-like protease or 3CL protease (nsp5), and RNA-dependent RNA polymerase (NSP12) and helicase (NSP13) (Chan et al. 2020). The other proteins encoded by the genome of the virus include structural proteins viz. spike glycoprotein, nucleocapsid, membrane and envelope proteins (Kumar et al. 2020). Following entry into the host cell, the viral RNA encodes two polyproteins (pp1a and pp1ab) which are subsequently cleaved into non-structured proteins (NSPs) by two viral proteases, viz. the main protease (3CLpro) and Papain-like protease (PLpro), resulting in the formation of replication-transcription complex (Li and Clercq 2020).
Once such viruses enter host cells, pattern recognition receptors (PRRs), including Toll-like receptors and retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), activate kinases which activate several transcription factors like interferon (IFN) regulatory factor-3 (IRF3), nuclear factor κB (NF-κB), etc. which lead to production of IFN. Ubiquitination and ISGylation are the primary signaling events which activate the various mediators and signaling molecules for the production of IFN (Devaraj et al. 2007; Thiel and Weber 2008; Perlman and Netland 2009; Zielecki et al. 2013). Several researchers reported that SARS-CoV can evade host innate immune responses by inhibiting production of IFN (see (Fung and Liu 2019) for details). The PLpro of SARS-CoV has been reported antagonize the production of IFN, and was found to possess deubiquitinase activity, and inactivate several key signal molecules involved in the generation of IFN (Sulea et al. 2005; Barretto et al. 2006; Frieman et al. 2009; Clementz et al. 2010). Recent studies revealed that the PLpro of SARS-CoV-2 preferentially strips off the ubiquitin-like interferon-stimulated gene 15 protein (ISG15), indicating de-ISGylation to be the preferred activity thereby attenuating type I INF response. It was further noted that inhibition of the PLpro maintained the INF pathway and reduced viral replication (Shin et al. 2020), which makes PLpro one of the most attractive drug targets against the virus (Dömling and Gao 2020).
The present study aims at identifying different compounds, including FDA approved drugs, drugs under clinical trials against different viruses or other pathogens, and natural products, which may potentially inhibit the PLpro of the SARS-CoV-2, using molecular modeling. The aim was thus to identify and suggest putative PLpro inhibitors which may be used for clinical trials on COVID-19 patients. So far, as of the time of writing this manuscript, three other such studies exist, one published and two in Preprint (Arya et al. 2020; Elfiky and Ibrahim 2020; Wu et al. 2020). However, these studies used homology modeling to predict the structure of the protease for the want of the actual structure of the PLpro of the SARS-CoV-2. The crystal structure of the protease has recently been determined and released on April 1, 2020. However, this structure does not contain any bound inhibitor, and as such the site of binding of the inhibitor was not available. X-ray diffraction structure of the PLpro with bound inhibitor GRL0617 was made on August 12, 2020. Thus, the present study is novel, and highly significant.
Methodology
The drug target
The drug target for the present study is the PLpro of the SARS-CoV-2. The three-dimensional structure of the protease was downloaded from the RCSB Protein Data Bank, bearing PDB id 7JRN (https://doi.org/10.2210/pdb7JRN/pdb). The structure was determined using X-ray diffraction, at a resolution of 2.48 Å, R-value free of 0.287, R-value work of 0.245 and R-value observed of 0.247. The protein was expressed in E. coli, and was deposited to the PDB on August 12, 2020 by Sacco and colleagues. The structure contains 2 chains (A and J) containing 315 amino acid residues each. One Zn2+ ion each is available with both the chains, and the PLpro inhibitor GRL0617 is also bound to each of the chains. Studies in SARS-CoV revealed that Zn2+ is essential for catalytic activity of the PLpro, whereby it acts as a cofactor [16,24. On the contrary, others have reported the ion to be an inhibitor of the protease activity of the enzyme (Han et al. 2005; Báez-Santos et al. 2015). Since the detail of the PDB structure is not yet published, it is not yet clear which Zn2+ ion(s) available with different chains of the SARS-CoV-2 PLpro structure is a cofactor and which one is an inhibitor.
The PDB structure was downloaded from the database in.pdb format. The stereological quality of the structure was assessed using PROCHECK module of the PDBSum server (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl). The Ramachandran plot analysis revealed that 88.3%, 11.2% and 0.5% amino acid residues fall in the most favoured, additionally allowed and generously allowed regions of the plot respectively, while no residue fell in the disallowed region (Supplementary material 1). Thus, the 3-dimensional structure was found to be a good model for molecular docking analysis.
The drugs
A total of 67 compounds were selected based on literature review (Table 1). Twenty eight compounds were suggested by Wu et al. (2020) from ZINC database based on molecular docking approaches, and were predicted to have SARS-CoV-2 PLpro inhibitory potential. Similarly, 16 compounds were suggested by Arya et al. (2020). Biltricide has been suggested by both these authors. Nine compounds were selected which are known inhibitors of PLpro of SARS-CoV, MERS-CoV or other related viruses. Kim et al. (2014) reported five compounds from the plant Psoralea corylifolia which were found to have PLpro inhibitory potentials. Park et al. (2017) suggested inhibitory potentials of 10 polyphenols from Broussonetia papyrifera which possess PLpro inhibitory potential. All of these compounds were included in the modeling analysis. In addition, we included hydroxychloroquine in our study, since chloroquine was found by Arya et al. (2020) to have potency in inhibiting SARS-CoV-2 PLpro, and the drug hydroxychloroquine is a more active derivative of the chloroquine. All the 3-dimensional structures of the compounds were downloaded from the NCBI PubChem compounds database (https://pubchem.ncbi.nlm.nih.gov/compound/) in.sdf format. Since the 3-dimensional structures of Iopromide, Telaprevir and Grazoprevir were not available in the database, their 2-dimensional conformers were downloaded and then 3-dimensional conformers were developed, followed by energy minimization. The properties of the ligands including molecular weight, octanol/water partition coefficient (logP), topological polar surface area (TPSA), and number of hydrogen bond donor (HBD) and acceptor groups (HBA) were obtained from the PubChem compounds database.
The molecular docking
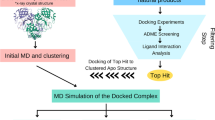
The structure of the drug target was loaded into the MoleGro Virtual docker 6.0 software (MVD). MolDock scoring function used in MVD is one of the most accurate molecular docking algorithms compared to several other such algorithms, and provides accuracy of 87% (Thomsen and Christensen 2006). One of the key scoring functions available in MVD is the Rerank score, which cross-validates the MolDock scores and identifies the most promising docking solution from the solutions obtained by the docking algorithm (Thomsen and Christensen 2006). The water molecules were removed from the workspace, and only chain A with its inhibitor and Zinc ion was included in the docking study. Taking the bound inhibitor GRL0617 as the reference ligand, the molecular docking was performed and the docking site was selected to be X: 8.37, Y: − 17.46 and Z: 31.28, including amino acids within a radius of 15 Å in the binding site. Scoring function was MolDock, and Grid resolution of 0.30 Å was selected. A total of 10 runs with 1500 iterations each were carried out. Using the above mentioned parameters, the docking was performed following standard procedures (Mazumder et al. 2019, 2020), to determine the affinities and geometries of binding of the compounds at the active site of the enzyme. The best pose of the ligands, in terms of MolDock score, was retained for further analysis.
Statistical analysis
To determine the properties of the ligands which are crucial for inhibition of the enzyme, we performed statistical correlation using MolDock score, Rerank score and Hydrogen bond score of the ligands with molecular weight, logP, HBD, HBA and TPSA, using Microsoft Office Excel 2007, and the Pearson’s correlation coefficient was determined. Before performing statistical analysis, the docking scores were converted to positive values.
Results
Binding of the drugs
Molecular docking using MVD determines the geometry of the ligands (drugs) while determining the free energies of binding. As such, the poses of binding of the compounds used in the present study were determined. While performing the docking, the bound inhibitor GRL0617 was also included to validate the docking. Interestingly, it was found that the compound binds to the same site of the enzyme in the same geometry (Fig. 1b), as that of its native bound state (Fig. 1a). This shows accuracy of the present modeling study. Further, the results demonstrate that all the ligands could effectively bind to the same active site. Although Zn2+ was included in the workspace, the ion does not show any interaction with the inhibitors and was away from this active site of PLpro. The docked poses of the 7 best inhibitors are shown in Fig. 1.
Docked poses of different ligands at the active site of SARS-CoV-2 PLpro. a The pose of the bound inhibitor as was available with the receptor; b docked poses of the same bound inhibitor, c Stallimycin, d Telaprevir, e Kazinol J, f Cefamandole, g Acetophenazine, h Sildenafil and i Boceprevir, as obtained following docking using MVD
Inhibition of the drug target
When a ligand or drug binds with the active site of an enzyme, it interferes with the binding of the substrate(s). Furthermore, the binding or docking energies indicate the amount of energy released when a ligand docks with the receptor, and thus predict the affinity of a ligand for the enzyme. In the present study, it was found that out of the 67 compounds studied, 26 compounds showed better inhibitory potentials compared to the bound inhibitor GRL0617 (Table 1). Among these compounds, the drug Stallimycin is the best inhibitor with highest (least negative) MolDock score (− 185.81 kcal/mol), and is followed by Telaprevir and Kazinol J. Stallimycin is also the best inhibitor of the enzyme among all the compounds suggested by Wu et al. (2020). However, Kazinol J was found to show the best Rerank score of − 130.46 kcal/mol, followed by Acetophenazine and Stallimycin (Table 1). Interestingly, among these compounds, Kazinol J showed the best hydrogen bond score of − 6.3682 kcal/mol, and is the best inhibit among the compounds suggested by Park et al. (2017). On the other hand, 4′-O-methylbavachalcone was found to be the best inhibitor among the compounds suggested by Kim et al. (2014).
Thus, the docking results indicate that all these suggested compounds have the potential to inhibit the PLpro of the SARS-CoV-2, albeit with different potentials. The chemical structures of the three best inhibitors, as revealed by the present study, are provided in Fig. 2. The inhibitory potential of Stallimycin, Telaprevir and Kazinol J were compared to the bound inhibitor (GRL0617), and were found to be 1.36-, 1.31- and 1.2-fold higher. The binding scores of the compounds which showed inhibitory potentials lesser than GRL0617 are given in Supplementary material 2.
Crucial properties of the ligands
The statistical correlation was performed to find the correlation coefficient and thereby to determine the crucial properties of the ligands which determine their binding affinities or docking scores. However, since the docking scores are negative values, and more negative or smaller the score better is the inhibition of the receptors, thus the docking scores were considered positive values for this analysis, following Mazumder et al. (2020). The analysis revealed that the molecular weights of the ligands are positively correlated to the MolDock score with coefficients of 0.622, while number of HBD groups present on the ligands was found to be positively correlated to the hydrogen bond score (coefficient 0.5015) (Table 2).
Discussion
Computational molecular docking is one of the most powerful tools in drug discovery research, and is frequently employed to identify drugs or lead molecules for their inhibitory potentials on known targets or enzymes (Mazumder et al. 2019, 2020). In the wake of drug repurposing against COVID-19, a large number of studies have been performed to identify known compounds which may be clinically tried for their effectiveness against different drug targets of the SARS-CoV-2. Of the different available drug targets of the virus, PLpro is one of the most attractive ones, since the enzyme has deubiquitination and deISGylation activities. Owing to this, it inactivates several signal molecules which need to be ubiquitinated and ISGylated to generate host immune response, and thus evades the innate immune responses (Fung and Liu 2019; Chen et al. 2014). Thus, inhibiting the enzyme alleviates the levels of INF and thereby the suppression of the host immune responses (Fig. 3) as well as attenuates replication of the virus (Shin et al. 2020).
Mechanism of PLpro inhibition-mediated inactivation of SARS-CoV-2. Ubiquitination and ISGylation are critical events in the innate immune responses against viruses. PLpro of SARS-CoV-2 helps the virus in evading host immunity by reversing these two critical steps. Thus, inhibitors against PLpro are hypothesized to counter this evasion mechanism of the virus. This might prevent the virus from replicating as well
The present modeling study employed molecular docking using computational tools to predict efficacy of different known compounds as well as FDA-approved drugs to target the PLpro of the SARS-CoV-2. The study screened 67 compounds for their effectiveness in inhibiting the target. So far, as of the time of writing this manuscript, and to the best of our knowledge, only three similar molecular modeling studies using SARS-CoV-2 PLpro exist, including two Preprints (Arya et al. 2020; Elfiky and Ibrahim 2020; Wu et al. 2020). However, these studies used Homology modeling based on the structure and sequences of SARS-CoV to determine the structure of the SARS-CoV-2 PLpro, and not the actual PLpro structure of the virus. This might be one of the reasons that although the studies used similar methodologies, the results in terms of the drugs they reported as inhibitors of PLpro were different, and there was only one such compound which was suggested by both Arya et al. (2020) and Wu et al. ( 2020). Thus, ours is the first study using the PLpro of the SARS-CoV-2, and is thus highly significant.
The present study revealed that Stallimycin is the best inhibitor of the SARS-CoV-2 PLpro, in terms of MolDock score. The compound was earlier suggested by Wu et al. (2020) using in silico approaches to be a potent inhibitor of the enzyme, and thus our finding is in line with the earlier work. The drug is an FDA-approved, oligopeptide antineoplastic antibiotic, and it was first isolated from Streptomyces distallicus. It has antiviral and antiprotozoal activities. Its effectiveness in inhibiting human papillomavirus has been reported earlier (Wetzler et al. 2011), and its analogues were found to have inhibitory activity against Trypanosoma burcei (Franco et al. 2020). However, these functions are attributed to its DNA-binding potential, while its antiviral potential by way of inhibiting PLpro of SARS-CoV-2 is a novel finding, and may be exploited for developing therapeutics against COVID-19. However, no clinical trial of the same on CODIV-19 is being done so far.
Among known viral protease inhibitors screened in the present study, Telaprevir, Grazoprevir and Boceprevir were found to be among the most potent inhibitors of the PLpro of SARS-CoV-2 (Table 1). Telaprevir is an orally active peptidomimetic drug that inhibits the protease of HCV (Vermehren and Sarrazin 2011). It was originally approved for the treatment of HCV, and is known to reduce viral replication (Jazwinski and Muir 2011) as well as facilitate IFN production (Meurs and Breiman 2007). Grazoprevir is a directly acting antiviral drug that inhibits the protease of the HCV (Keating 2016). Telaprevir, Boceprevir and Grazoprevir are approved inhibitors of PLpro of HCV (Gonzalez-Grande et al. 2016; Saleh et al. 2014; Tong et al. 2012; Sarrazin et al. 2012). Other notable compounds that were found to inhibit the SARS-CoV-2 PLpro include GRL0617, Mycophenolic acid and GRL0667 (Table 1). In vitro study using Vero E6 cells revealed that GRL0617 inhibits the SARS-CoV replication with IC50 of 15 µM, and had no cytotoxicity (Ratia et al. 2008). Similarly, Mycophenolic acid, derived from Penicillium stoloniferum, inhibits SARS-CoV PLpro (Lee et al. 2015). GRL-0667, GRL-0617 and Mycophenolic acid are under clinical trial against SARS-CoV PLpro. Since the protease of SARS-CoV-2 bears similarities with that of SARS-CoV, and in view of the present finding, it is surmised that these compounds would turn out to be effective against SARS-CoV-2 as well. Thus, it is suggested that in vitro studies as well as clinical trials are initiated using these compounds against SARS-CoV-2.
In addition, the polyphenols suggested by Kim et al. (Kim et al. 2014) against SARS-CoV were also found to be potent inhibitors of SARS-CoV-2 PLpro (Table 1; Supplementary material 2). Among them, 4′-O-methylbavachalcone was found to be the best inhibitor, followed by Corylifol A (Table 1). The compounds are found in the seeds of the traditional Chinese and Indian medicinal herb P. corylifolia (Kim et al. 2014). The seed of this plant is edible, and are used against several ailments including skin diseases, such as psoriasis and leukoderma, and leprosy, vitiligo, asthma, ulcers and kidney disorders (see (Khushboo et al. 2010; Chopra et al. 2013; Koul et al. 2018) for details). The polyphenolic compounds found in the plant B. papyrifera (paper mulberry) were reported to be potent in inhibiting the PLpro of SARS-CoV (Arya et al. 2020). Further, all the 10 polyphenols isolated from B. papyrifera, and reported to be effective against PLpro of SARS-CoV and MERS-CoV by Park et al. (Arya et al. 2020), were also found to be potent in inhibiting the PLpro of SARS-CoV-2 (Table 1). Among these, Kazinol J was found to be the most potent in the present study. B. papyrifera is a deciduous tree distributed across Asia–Pacific and the USA. The fruit, leaves and roots of the plant are edible, and have been in use in Chinese traditional medicine against several ailments including prostatitis, impotence and ophthalmic disorders (Sun et al. 2012). These results indicate that the polyphenols may potentially be inhibitors of PLpro, and thus different polyphenols including those obtained from tea, turmeric, etc. may be tried for drug discovery research.
Conclusion
The PLpro of the SARS-CoV-2 is one of the most vital drug targets to control replication of the virus inside host. Studies in other related CoVs, including SARS and MERS, and recent findings on SARS-CoV-2 revealed that inhibition of the enzyme attenuates the processing of the viral polyprotein as well as alleviates the suppression of the host innate immune responses. The present study evaluates 68 different drugs and compounds which are known inhibitors of PLpro of related viruses, re-purposed drugs or other compounds known to have PLpro inhibiting potentials. While Stallimycin was found to be the best inhibitor of the enzyme, known inhibitors including Telaprevir, Grazoprevir and Boceprevir were highly potent in inhibiting the enzyme. In addition, several polyphenols derived from plants and reported to inhibit the enzyme of SARS and MERS were also found to be potent inhibitors, among which Kazinol J showed promising results. Based on the findings, we suggest that clinical trials be initiated with these inhibitors. Since less focus has so far been given on this crucial aspect of PLpro inhibition in containing the COVID-19, the present study is novel and has therapeutic implications in containing the pandemic.
References
Arya R, Das A, Prashar V, Kumar M (2020) Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs. ChemRxiv. Preprints. https://doi.org/10.26434/chemrxiv.11860011.v2
Baker EN (2020) Visualizing an unseen enemy; mobilizing structural biology to counter COVID-19. Acta Cryst F76:158–159. https://doi.org/10.1107/S2053230X20004847
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC (2005) The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79:15189–15198
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC (2006) Deubiquitinating activity of the SARS-CoV papain-like protease. Adv Exp Med Biol 581:37–41
Báez-Santos YM, St John SE, Mesecar AD (2015) The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir Res 115:21–38. https://doi.org/10.1016/j.antiviral.2014.12.015
Chan JF, Kok KH, Zhu Z et al (2020) Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9(1):221–236. https://doi.org/10.1080/22221751.2020.1719902
Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z (2014) SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 5(5):369–381. https://doi.org/10.1007/s13238-014-0026-3
Chen N, Zhou M, Dong X et al (2020) Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. https://doi.org/10.1016/S0140-6736(20)30211-7
Chopra B, Dhingra AK, LalDhar K (2013) Psoralea corylifolia L. (Buguchi)—Folklore to modern evidence: review. Fitoterapia 90:44–56. https://doi.org/10.1016/j.fitote.2013.06.016
Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, BaezSantos YM, Wang J, Takayama J, Ghosh AK et al (2010) Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629
Cunningham AC, Goh HP, Koh D (2020) Treatment of COVID-19: old tricks for new challenges. Crit Care 24:91. https://doi.org/10.1186/s13054-020-2818-6
Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC et al (2007) Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221
Dömling A, Gao L (2020) Chemistry and Biology of SARS-CoV-2. Chem 6:1283–1295. https://doi.org/10.1016/j.chempr.2020.04.023
Elfiky A, Ibrahim NS (2020) Anti-SARS and anti-HCV drugs repurposing against the Papain-like protease of the newly emerged coronavirus (2019-nCoV). Res Square. Preprints. https://doi.org/10.21203/rs.2.23280/v1
Franco J, Scarone L, Comini MA (2020) Novel distamycin analogues that block the cell cycle of African trypanosomes with high selectivity and potency. Eur J Med Chem 189:112043. https://doi.org/10.1016/j.ejmech.2020.112043
Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS (2009) Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83:6689–6705
Fung TS, Liu DX (2019) Human coronavirus: host-pathogen interaction. Annu Rev Microbiol 73:529–557
Gonzalez-Grande R, Jimenez-Perez M, Gonzalez Arjona C, Mostazo TJ (2016) New approaches in the treatment of hepatitis C. World J Gastroenterol 22(4):1421–1432
Guo D (2020) Old weapon for new enemy: drug repurposing for treatment of newly emerging viral diseases. Virol Sin. https://doi.org/10.1007/s12250-020-00204-7
Han YS, Chang GG, Juo CG, Lee HJ, Yeh SH, Hsu JT, Chen X (2005) Papainlike protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry 44:10349–10359
Jazwinski AB, Muir AJ (2011) Direct-acting antiviral medications for chronic hepatitis C virus infection. Gastroenterol Hepatol (N Y) 7(3):154–162
Keating GM (2016) Elbasvir/Grazoprevir: first global approval. Drugs 76(5):617–624. https://doi.org/10.1007/s40265-016-0558-3
Khushboo PS, Jadhav VM, Kadam VJ, Sathe NS (2010) Psoralea corylifolia Linn.—“Kushtanashini”. Pharmacognosy Reviews 4(7), 69–76. https://doi.org/10.4103/0973-7847.65331
Kim DW, Seo KH, Curtis-Long MJ, Oh KY, Oh JW, Cho JK, Lee KH, Park KH (2014) Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzyme Inhib Med Chem 29(1):59–63. https://doi.org/10.3109/14756366.2012.753591
Koul B, Taak P, Kumar A, Kumar A, Sanyal I (2018) Genus Psoralea: a review of the traditional and modern uses, phytochemistry and pharmacology. J Ethnopharmacol 232:201–226. https://doi.org/10.1016/j.jep.2018.11.036
Kumar S, Nyodu R, Maurya VK et al (2020) Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). In: Saxena S. (eds) Coronavirus Disease 2019 (COVID-19). Medical Virology: From Pathogenesis to Disease Control. Springer, Singapore. https://doi.org/10.1007/978-981-15-4814-7_3
Lee H, Lei H, Santarsiero BD et al (2015) Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem Biol 10(6):1456–1465
Li G, De Clercq E (2020) Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discovery 19:149–150
Lu R, Zhao X, Li J et al (2020b) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. https://doi.org/10.1016/S0140-6736(20)30251-8
Lu H, Stratton CW, Tang Y (2020a) Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol 25678
Mazumder MK, Borah A, Choudhury S (2020) Inhibitory potential of plant secondary metabolites on anti-Parkinsonian drug targets: relevance to pathophysiology, and motor and non-motor behavioural abnormalities. Med Hypotheses 137:109544. https://doi.org/10.1016/j.mehy.2019.109544
Mazumder MK, Choudhury S, Borah A (2019) An in silico investigation on the inhibitory potential of the constituents of Pomegranate juice on antioxidant defense mechanism: relevance to neurodegenerative diseases. IBRO Rep 6:153–159
Meurs EF, Breiman A (2007) The interferon inducing pathways and the hepatitis C virus. World J Gastroenterol 13(17):2446–2454
Park JY, Yuk HJ, Ryu HW, Lim SH, Kim KS, Park KH, Ryu YB, Lee WS (2017) Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J Enzyme Inhib Med Chem 32(1):504–512. https://doi.org/10.1080/14756366.2016.1265519
Perlman S, Netland J (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450
Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Meseca AD (2008) A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. PNAS 105(42):16119–16124
Saleh NA, Elfiky AA, Ezat AA, Elshemey WM, Ibrahim M (2014) The electronic and quantitative structure activity relationship properties of modified telaprevir compounds as HCV NS3 protease inhibitors. J Comput Theor Nanosci 11(2):544–548
Sarrazin C, Hézode C, Zeuzem S, Pawlotsky J-M (2012) Antiviral strategies in hepatitis C virus infection. J Hepatol 56:S88–S100
Shin D, Mukherjee R, Grewe D et al (2020) Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. https://doi.org/10.1038/s41586-020-2601-5
Sulea T, Lindner HA, Purisima EO, Menard R (2005) Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J Virol 79:4550–4551
Sun J, Liu S, Zhang C, Yu L, Bi J, Zhu F, Yang Q (2012) Chemical composition and antioxidant activities of Broussonetia papyrifera Fruits. PLoS ONE 7(2):e32021. https://doi.org/10.1371/journal.pone.0032021
Thiel V, Weber F (2008) Interferon and cytokine responses to SARScoronavirus infection. Cytokine Growth Factor Rev 19:121–132
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321. https://doi.org/10.1021/jm051197e
Tong J, Wang YW, Lu YA (2012) New developments in small molecular compounds for anti-hepatitis C virus (HCV) therapy. J Zhejiang Univ Sci B 13(1):56–82
Vermehren J, Sarrazin C (2011) New HCV therapies on the horizon. Clin Microbiol Infect 17(2):122–134
Wetzler DE, Comin MJ, Krajewski K, Gallo M (2011) New human papilloma virus E2 transcription factor mimics: a tripyrrole-peptide conjugate with tight and specific DNA-recognition. PLoS ONE 6(7):e22409. https://doi.org/10.1371/journal.pone.0022409
Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. https://doi.org/10.1016/j.apsb.2020.02.008
Zielecki F, Weber M, Eickmann M, Spiegelberg L, Zaki AM, Matrosovich M, Becker S, Weber F (2013) Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol 87:5300–5304
Funding
The present study has not been funded by any organization.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical disclosure
The present study uses no animal model or human subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary material 1:
Ramachandran plot and the related statistics of the PDB structure used in the study. (PDF 14 kb)
Supplementary material 2:
Docking scores of the compounds which showed lesser potentials compared to GRL0617 in inhibiting SARS-CoV-2 PLpro. (DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Choudhury, S., Moulick, D., Borah, A. et al. In search of drugs to alleviate suppression of the host's innate immune responses against SARS-CoV-2 using a molecular modeling approach. In Silico Pharmacol. 9, 26 (2021). https://doi.org/10.1007/s40203-021-00085-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-021-00085-y