Abstract
High-energy diffraction microscopy (HEDM) in-situ mechanical testing experiments offer unique insight into the evolving deformation state within polycrystalline materials. These experiments rely on a sophisticated analysis of the diffraction data to instantiate a 3D reconstruction of grains and other microstructural features associated with the test volume. For microstructures of engineering alloys that are highly twinned and contain numerous features around the estimated spatial resolution of HEDM reconstructions, the accuracy of the reconstructed microstructure is not known. In this study, we address this uncertainty by characterizing the same HEDM sample volume using destructive serial sectioning (SS) that has higher spatial resolution. The SS experiment was performed on an Inconel 625 alloy sample that had undergone HEDM in-situ mechanical testing to a small amount of plastic strain (~ 0.7%), which was part of the Air Force Research Laboratory Additive Manufacturing (AM) Modeling Series. A custom-built automated multi-modal SS system was used to characterize the entire test volume, with a spatial resolution of approximately 1 µm. Epi-illumination optical microscopy images, backscattered electron images, and electron backscattered diffraction maps were collected on every section. All three data modes were utilized and custom data fusion protocols were developed for 3D reconstruction of the test volume. The grain data were homogenized and downsampled to 2 µm as input for Challenge 4 of the AM Modeling Series, which is available at the Materials Data Facility repository.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
This data descriptor article describes a 3D microstructural reconstruction experiment associated with the Air Force Research Laboratory (AFRL) additive manufacturing (AM) modeling series [1]. The AM Modeling Series consisted of a complementary set of four challenge problems that collectively examined the capability of modeling and simulation toolsets to quantitatively predict aspects of structure or property heterogeneity within metal AM test articles. The 3D data volume that is the subject of this article is associated with the 4th Challenge that assessed the accuracy of image-based simulations that utilize an explicit 3D reconstruction of the grain microstructure to predict both the aggregate stress–strain behavior and localized response in selected grains.
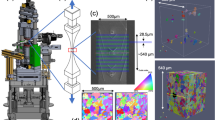
All of the AM Modeling Challenge Problems utilized Inconel 625 samples printed on an EOS M280 Laser Powder Bed Fusion system (LPBF), using commercially available gas atomized powder. The bulk sample material in this specific challenge went through a stress relief heat treatment, hot isostatic pressing and additional heat treatment. The test sample was prepared by wire electrical discharge machining (EDM), and no further surface treatments or machining was performed prior to mechanical testing. A schematic of the HEDM tensile sample is shown in Fig. 1.
Schematic of a HEDM tensile sample, with the blue colored rectangle approximately representing the spatial position of the volume-of-interest (VOI). The SEM image on the right side of this figure highlights the fiducial markings on the surface of the HEDM sample. The gold-colored arrows indicate the position of the three HEDM gold markers that denote the end points and middle of the VOI, the green-colored arrows indicate multiple FIB-deposited platinum fiducial lines within the VOI, and the red-colored arrow shows the location of the FIB milled trench that assisted aggressive step polishing prior to the start of the SS experiment
This challenge problem utilized non-destructive HEDM measurements [2] performed at the Advanced Photon Source at Argonne National Laboratory to quantify the local microstructural orientation and strain tensor within individual grains at selected states during the in-situ mechanical test. As a result, the sample characterized in this SS experiment has been plastically deformed to a small amount, εp ~ 0.7%. For brevity, we refer the reader to an associated data descriptor article in this journal that details the HEDM experiment associated with Challenge 4 [3]. In addition, the collective input data associated with this challenge are hosted by the Materials Data Facility [4, 5] and the Digital Object Identifier to the Challenge 4 input data is listed here [6], which includes additional supplementary information.
One aspect of this experiment that was the subject of significant deliberation by the authors prior to designing the experimental workflow was the desired spatial resolution of the SS data set. A recent study of the spatial precision of near-field HEDM reconstructions by some of the authors report a value of approximately 2 µm [7]. It is important to note that this value was determined by analysis of the spatial position of grain boundaries for equiaxed grains with lateral dimensions much larger than the reported resolution and minimal intragranular orientation gradients, which is unlike the present study. The authors anticipated that the presence of small grains with dimensions on the order of the spatial resolution was likely to be improperly resolved. Therefore, the desired spatial resolution of the SS reconstruction was to match the reported resolution of HEDM near-field reconstructions, that is, a 2 µm voxel resolution. One practical issue with long-term SS experiments is that artifacts or other unexpected changes can cause degradation or loss of information. The destructive nature of the SS experiment eliminates any chance for recollection once the next section is started. Thus, the SS data were collected at a voxel resolution of approximately 1 µm in order to minimize the effect of missing or corrupted data from the automated SS experiment, which was ultimately homogenized and downsampled to 2 µm for the Challenge 4 input data.
Sample Preparation and Mounting
Relative to other SS experiments performed by the authors, considerable effort was expended to ensure that the relatively fragile tensile sample was optimally prepared for mechanical polishing SS while minimizing the potential for accidental damage, and also ensured that the volume-of-interest (VOI) could be readily identified after embedding the sample in an opaque, SEM-compatible mount. The following section describes the multi-step sample procedure used for this experiment.
After the in-situ mechanical testing experiment, plasma-focused ion beam (FIB) milling was used to separate the tensile sample into two pieces, such that the VOI was approximately 1 mm from the end of one of the pieces. Next, multiple linear features were added to the sample surface via liquid metal ion source FIB microscopy, to aid in assessment of the depth of SS from analysis of the as-polished surface. It is critically important in a SS experiment to monitor the depth that a specific VOI resides below the surface of an opaque metallurgical mount, as unintended polishing through the VOI is an irrecoverable failure. Three cuboidal gold markers (~ 40 µm edge length) were previously placed using a combination of micromanipulation, FIB-based milling and deposition methods on the tensile sample prior to the in-situ experiment to denote the VOI [8]. The three markers correspond to the middle and end-points of the VOI that was characterized by both near-field and far-field HEDM. To supplement these features, multiple inclined platinum lines were FIB-deposited on the sample surface as shown in Fig. 1, which are readily observable in both optical and backscatter SEM images of the as-polished SS sample surface. Deposition of these fiducial marker lines avoided any alteration or local material removal at the test sample surface, and the presence of multiple lines along the length of the sample enabled an unambiguous assessment of the approximate sectioning depth from either a single image of the cross-section surface or a 3D reconstruction.
For the region of the sample that would be removed by polishing prior to reaching the VOI, a linear trench was FIB milled into the surface at an inclined angle to the tensile axis over a relatively large distance (~ 0.75 mm along the sectioning direction). Similar to the Pt lines, this feature assisted with assessment of the location of the VOI relative to the current SS surface. Since this material was not used in reconstruction of the gage volume, milling was selected over Pt-deposition as this process could be more efficiently completed.
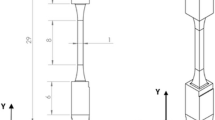
After FIB-machining and marking, the sample was embedded in an electrically conductive thermoset mount to facilitate the use of mechanical polishing methods to SS the relatively small HEDM test sample—0.5 × 0.5 mm cross-section—and also employ a high-current probe for Electron Backscatter Diffraction mapping. A custom support structure made from a titanium alloy was designed and manufactured to assist with both centering and axial alignment of the tensile sample within the mounting press, and to create a millimeter-sized gap between the mounting press ram and the tensile sample to avoid deforming the sample during pressurized hot consolidation. Representative images of the custom titanium support structure are shown in Fig. 2. While it was not initially considered, the incorporation of a higher hardness alloy in the materialographic mount also slowed the rate of material removal via polishing, which may have contributed to the stability of the SS removal rate throughout the long-duration experiment.
a CAD model of a custom titanium alloy sample holder (gray) that was used to optimize the centering and depth of the tensile sample (red) within the 1½” dia. cylindrical metallographic hot mount. b Side view of the CAD model and photograph of the sample holder that highlights the recessed placement of the end of the tensile sample and the surface of the sample holder
The tensile sample and the titanium support structure were hot mounted in a 1.5″ dia. cylinder using Struers Polyfast compound and a Struers CitoPress-20 using manufacturer prescribed conditions for sensitive samples, where no pressure is used during the initial two-minute heating segment. During powder filling of the mounting cylinder, a vibrating engraving tool was placed in direct contact with the cylinder to help to maximize the packing of powder into the crevasses of the titanium support structure prior to consolidation. After mounting, radiographic x-ray imaging was performed to ensure that the sample was in the desired condition prior to SS. The radiographic images showed that the tensile sample axis was slightly misaligned relative to vertical surfaces of the titanium holder (and correspondingly the anticipated SS surface). Image analysis of two radiographic images collected at orthogonal views estimated that the sample was inclined by 0.6° and 1.5°, respectively, to the titanium support structure.
Prior to SS, the outer rim of the thermoset mount was machined using a lathe to create a ledge surrounding the SS surface. The purpose of this offset ledge is to provide a convenient reference surface that is not altered during the SS experiment, which can be used to perform differential depth measurements using a variety of methods (focus values from optical microscopes, mechanical or laser-based profilometers, or a dial indicator, as discussed in [9]). Lastly, the sample was permanently affixed to a RoboMet.3D™ stainless steel sample fixture using a SEM-compatible epoxy (Epo-Tek Silver Conductive Epoxy, Ted Pella). Figure 3 shows images of the materialographic sample mount attached to the stainless steel fixture, which was taken after completion of the SS experiment.
Serial Sectioning (SS)
SS was performed using a custom-built system at AFRL—colloquially referred to as LEROY—that is capable of fully automated multi-step materialographic sample preparation and multi-modal microscopy workflows, including the collection of scanning electron microscopy data [10, 11]. Materialographic sample preparation and cleaning processes within the LEROY system are performed by a significantly modified, second-generation RoboMet.3D™ outfitted with 300 mm dia. polishing pads, which can achieve a wide range of SS thickness values while potentially finishing with nanometer-scale abrasives to create low-damage surfaces that are sufficient for EBSD-based mapping. The RoboMet.3D™ within the LEROY system includes a Zeiss AxioVert inverted optical microscope that can be used for bright field, dark field and polarized light microscopy collection. The LEROY system also incorporates a Tescan Vega 3 XMH LaB6 SEM with automated load lock exchange and Bruker Quantax e-Flash 1000 EBSD detector, and two additional Zeiss optical microscopes (Zeiss AxioImager.Z2 and AxioObserver), allowing the user to select and optimize the most appropriate modes of microscopy to include in the SS experiment for subsequent analysis.
The mechanical polishing workflow of the experiment consisted of two material removal steps that were performed by the RoboMet.3D™, using a constant force of ~ 24.5 N. The primary material removal step utilized 1 μm polycrystalline diamond abrasive in a water-based suspension (Allied High Tech) diluted with a concurrent drip of distilled water on a woven acetate polishing pad (DAC, Struers) for approximately 5 min at a rotational speed of 100 rpm. The second polishing step utilized 40 nm-scale colloidal silica suspension (non-stick rinsable, Allied High Tech.) diluted on a non-woven porous polyurethane polishing pad (Final A, Allied High Tech.) with a concurrent drip of distilled water for 2 min at a rotational speed of 100 rpm. After each of these two steps, a long-nap cloth (Final Pol, Allied High Tech.) with flowing tap water was used to clean the sample surface of polishing media (30 s at a rotational speed of 75 rpm) to minimize grit cross-contamination. Ultrasonic cleaning using pure Ethanol as the immersion fluid was performed prior to microscopic data collection, using a sequence of multiple beakers and nitrogen gas drying to minimize contaminants on the freshly exposed SS surface. Note that cube-corner microhardness indents were intermittently introduced to the SS surface (via MTS NanoXP Nanoindentation system, 9 N indentation load) throughout the experiment to the titanium support structure that surrounded the tensile sample, as a contingency protocol to quantify SS material removal rates [12], but were not used as optical microscope focus value measurements were deemed sufficient.
The microscopy workflow consisted of three types of data collection: bright-field optical montage images, EBSD maps, and backscatter SEM images, which are described in the remainder of this section. Bright-field optical images were included for two purposes. First, these were used as a data mode that allowed for robust registration of the volumetric stack of serial section microscopy data. Systematic errors in section-to-section alignment can occur when only one dominant feature is present in each section, e.g., the tensile sample cross-section, and a metric that maximizes the overlap of this dominant feature is used for stack registration [13]. This was of particular concern as the radiographic imaging of the metallographic mount indicated that the longitudinal axis of the tensile sample was slightly misaligned relative to the SS surface normal. To avoid this problem, the use of materialographic mount material that contained carbon filler particles, combined with the use of large-area montage image data collection that created images with thousands of randomly oriented particles, enabled the use of the SIFT algorithm [14] to calculate the optimal registration. Second, a common method to measure the depth of the SS surface is by recording the value of the optical microscope focus motor position [9], which can be effective when both high numerical aperture objective lenses (which have a narrow depth of field) are used in conjunction with automated focusing algorithms.
A Zeiss AxioVert 200 M inverted optical microscope outfitted with a AxioCam MRc5 camera was used to collect a bright-field 12-bit grayscale images from the SS surface, using a 20× objective lens (Epiplan, 0.4 NA) and a 1.0× optivar, resulting in an image pixel dimension of 0.52 µm. For the SS Region-Of-Interest (ROI), a 144 tile (12 × 12) montage using a single focus point and ~ 50% image overlap was collected at each section, which covered an area of approximately 6.2 × 4.6 mm. This relatively large ROI (compared to the tensile sample cross section) also included subregions of the titanium support structure with microhardness indents for contingency stack registration and depth measurements. A highly cropped region of the optical montage image data (highlighting just the tensile sample cross section) is shown in Fig. 4a. Three additional 3 × 3 montages were collected—one at the center of the ROI and two at opposing ends of the sample within the offset ledge—using the same microscope conditions but with autofocusing at every third tile, to allow for differential SS depth measurements.
Representative microscopy data from the SS experiment, where all three panels show data from the same section. a Highly cropped subregion of an optical montage image, highlighting the tensile sample cross-section and random microstructure of the materialographic mount. b EBSD map representing Inverse Pole Figure (IPF) coloring associated with the z-axis (out of plane) orientation, along with the Face-Centered Cubic IPF color legend c BSE image generated by combination of multiple images as discussed in the accompanying text, where the Pt fiducial markings are readily visible as bright features in the upper portion of the image
Electron Backscatter Diffraction maps were a key data mode for this experiment in order to provide a spatially resolved micron-scale characterization of the local crystal orientation. A single 600 × 600 pixel map with 1 µm pixel dimensions was collected at each section, as shown in Fig. 4b. These maps were collected at 70° stage tilt, using a 20 kV accelerating voltage and a high (5–13 nA) probe current, and 8X binning (80 × 60 EBSD patterns), enabling the Bruker detector to operate at 550 points per second, resulting in map collection times of approximately 11 min. Note that while the individual EBSD patterns from the tensile sample contained clearly defined Kikuchi bands, there was often a few percent of pixels that were not initially indexed by the Bruker Esprit 1.9.4 software, therefore both the original EBSD patterns (*.bcf) and the indexed data (*.ctf) were saved at each section to enable dictionary-based reindexing analysis [15]. Saving all of the EBSD patterns significantly increased the total amount of disk storage space for the experiment, as each *.bcf file was approximately 2 GB.
It is known that geometric distortions can commonly occur in EBSD data scans, which are accentuated by the large tilt of the sample surface relative to the incident electron beam [16]. As will be discussed in the following section, a data fusion approach was employed to address both geometrical distortions and stack registration by a non-linear spatial transform of the raw EBSD data to the registered stack of optical microscopy data. Initial attempts to register the EBSD maps directly to the optical image failed, as there were few common features in both data modes. Therefore, backscattered electron (BSE) images at normal incidence (0° stage tilt) were also collected at each section, as an intermediary data type to facilitate EBSD stack registration. As shown in Fig. 4c, the BSE images contain significant electron channeling contrast that correlated to the crystalline grain structure, and this contrast feature was used to register EBSD maps to the corresponding BSE images. Moreover, numerous micrometer-scale precipitates and pores within the tensile sample were also observable in both the optical and BSE images, which enabled registration of these data modes. Occasional and random local intensity fluctuations of the BSE signal were observed during image collection, thus five 1024 × 1024 pixel images with 0.59 µm pixel dimensions with 16-bit grayscale depth were collected at each section, using an accelerating voltage of 20 kV and a probe current of ~ 1 nA, and resulted in a collection time of approximately 3 min. These images were fused post-collection into a single image that eliminated the intensity fluctuations, using Fiji [17] to perform SIFT registration of the stack of 5 images followed by a z-projection median filter.
Considering all of the aforementioned steps of the SS experiment, the total cycle time for one section was approximately 63 min, which includes additional activities such as robotic manipulation within and between the various subsystems (e.g., polishing pad exchanges for RoboMet.3D, exchange of the sample into and out of the SEM via a load lock), and real-time optimization of key SEM acquisition parameters (e.g., working distance, focus, and brightness/contrast). The cumulative experiment consisted of 1000 sections that completely encompassed the volume denoted by the two outer gold markers, although only a subset of the 1000 sections was required for the Challenge 4 data. The initial 900 sections were collected in a 49-day span before a SEM vacuum pump failure occurred, which resulted in a few week delay before the final 100 sections were collected.
Figure 5a shows a plot of the calculated depth of the SS surface from optical microscope focus values as a function of section number for the experiment, and an ordinary least squares linear regression fit to this data. These material removal data were generated from averaging the microscope focus values of the aforementioned “additional 3 × 3 montages” that contain 3 autofocus operations per montage. The residuals for the linear regression fit are shown in Fig. 5b, and the authors note that occasional outlier values are present in the data, which were included in the calculation of the linear fit. For 3D reconstruction of the SS data, the authors selected to use a constant value for the voxel depth of 0.97 µm per section for the first 900 sections of the data set, and this selection is supported by the coefficient of determination (R2) value of 0.9998 for the linear fit. The choice of a constant depth value also greatly simplified some aspects of the reconstruction workflow. The residual errors to the linear fit in Fig. 5b are not entirely randomly distributed, and therefore efforts to calculate non-uniform (section-to-section) depth values will likely enhance the accuracy of subsequent 3D reconstructions, but have not been performed to date.
a Plot of cumulative material removed during the SS experiment relative to the serial section number (section numbers range from 384 to 1383). The material removal data were linear least squares fit (shown as a solid red line) for sections 384 to 1283, resulting in an average serial sectioning rate of 0.97 µm per section. The residuals from the linear fit are shown in (b). Note that the subvolume of data used for Challenge 4 approximately encompassed sections 410–1100
Registration and Fusion of SS Data
From the microscopy data shown in the previous section, one can observe that each data modality offered unique information from the SS experiment. In order to collectively utilize this information, the data from each modality had to be spatially registered to a common reference frame, which was accomplished using the workflow pictorially represented in Fig. 6. First, the stack of optical images were aligned and registered to each other. Once this was accomplished, the stack registered optical images were treated as the spatial ground truth upon which all the other modalities were registered on a section-by-section approach. Next, each BSE image was spatially registered to its corresponding section optical image, which was followed by spatially registering each EBSD map to the corresponding transformed BSE image. Details of the data registration and fusion process are described in the remainder of this section.
Pictorial representation of the workflow to register and fuse the three microscopy data modalities—bright field optical images, BSE images and EBSD maps. The optical images were used for accurate stack registration of the SS data, the BSE images were used as intermediary data to register the EBSD data modes to the optical image stack and allowed for simple segmentation of internal porosity, and the EBSD maps provided local, quantitative crystallographic information
The initial step in the SS data registration and fusion workflow was to assemble and fuse each section’s optical image tiles into montages, which were subsequently registered as an image stack to create a 3D data volume. First, the individual optical image tiles were corrected for illumination non-uniformities (a.k.a. flatfielding, shading correction) using custom python scripts to calculate and minimize this artifact. The flatfielded image tiles were spatially arranged and fused into a montage using the “Grid/Collection Stitching Plug-In” [18] in Fiji, with sub-pixel interpolation and linear blending enabled. Spatial registration of the stack of montage images was performed using the “Register Virtual Stack Slices” plug-in in Fiji [17], utilizing default parameters, save for selection of a “Rigid” expected transformation (allowing both translation and rotation), and the “interpolate” option enabled. After registration and transformation of the optical image stack, a subregion was cropped that was manually centered on the tensile sample, covering an area of approximately 0.65 × 0.65 mm (1156 × 1156 pixels), which facilitated section-to-section matching with the other data modalities. Visual examination of the registered optical microscopy data volume did not indicate the presence of a systematic bias in stack alignment.
The BSE images were registered to the optical images using custom python scripts. Prior to the registration step, the optical images were first normalized to enhance the image contrast within the tensile sample cross-section of the porosity and precipitates from the matrix, while concurrently eliminating image contrast within the surrounding mount material. The intensity values of the normalization were selected by manual inspection of the optical image histogram. Figure 6a shows an optical image before normalization, Fig. 6d shows the same optical image after normalization, and Fig. 6b shows the corresponding BSE image for the same section. The normalization step emphasized the common features in both the BSE and optical images, enhancing the likelihood of calculating a satisfactory alignment using an optimization algorithm.
The ITK multi-resolution image registration framework [19] was used to register the BSE images to the optical images. The multi-step registration starts with low resolution images, obtains a coarse alignment, and then progressively increases the resolution to optimize the alignment, using the previous lower-resolution result as an initial condition. Input parameters for the ITK transform included the following selections: b-spline for the transform, mutual information for the similarity metric [20], L-BFGS-B for the optimizer, and nearest neighbor interpolation. The BSpline mesh size was 1 × 1 and the order of the transformation was 2. For the other components, the default parameters were used. After transformation, the BSE images were contrast normalized using the “HistogramMatchingImageFilter” in SimpleITK. The first BSE image of the SS stack was selected as the target, and the histograms for all subsequent images were adjusted to match the target. The number of match points was set to 4 and all other parameters were set to the default values. This normalization step improved the consistency of grayscale values across the entire stack, which was useful for both visualization of the 3D data, and also for the final data registration task of alignment of the EBSD maps to the transformed BSE image stack.
Prior to this final registration step, the BSE images were downsampled from their original resolution of 1024 × 1024 pixels to 600 × 600 pixels, in order to match the pixel resolution of the EBSD maps. Rescaling the BSE images prior to performing the image registration improved the success rate of the optimization algorithm in determining a valid solution, and note that rescaling was not needed in the prior registration task as the BSE and OM images had similar pixel dimensions. In addition, the stack of EBSD data was reindexed by dictionary indexing [15], which was performed by Marc De Graef and Michael Jackson, and the reindexed maps were used in the remainder of the data fusion workflow. The reindexed EBSD maps were also converted to a monochrome image to facilitate the registration process, by simple grayscale conversion of one of the Inverse Pole Figure (IPF) color image outputs to an 8-bit grayscale image using the DREAM.3D filters “Generate IPF Colors” and “Color to Grayscale.” Spurious EBSD map values (correlated to data associated with the materialographic mount material) were also removed by identifying the largest unindexed region from the original EBSD map using the DREAM.3D filter “Isolate Largest Region (Identify Sample).” The ITK image registration framework was also used for registering the EBSD maps to the BSE images, using the same selections and values as described previously, save for the BSpline mesh size was 3 × 3. The parameter values used for data registration were determined by qualitative visual examination of the post-registered data, and a representative semi-transparent data overlay is shown in Fig. 6i to demonstrate the quality of data alignment. The optimal transformation found on a section-by-section basis for each map, and this transformation was applied to each of the EBSD maps in the *.ctf file format using a custom python script and nearest neighbor interpolation.
After registration of the three data types, the data were fused into a new *.ctf file using additional processing steps in DREAM.3D. First, the spatially transformed BSE images were segmented into 5 phases, using manual selection of multiple global thresholds for the grayscale values. Here, Phase 1 was the IN625 metallic tensile sample, Phase 2 was the internal porosity within the tensile sample, Phase 4 was the gold markers used to denote the HEDM VOI, Phase 5 was the Pt fiducials applied to assist with the SS experiment, and Phase 6 was materialographic mount material. A representative image of the segmented BSE data is shown in Fig. 6f. The authors note that micrometer-scale precipitates within the tensile sample were not segmented, and instead treated these features as part of the metal phase, as an adequate segmentation strategy was not developed (these were initially identified as Phase 3). Next, a new ‘fused’ *.ctf file was created that combined the phase information from the spatially transformed and segmented BSE images and with the local Euler orientation and other associated pattern values from the spatially transformed EBSD maps. For data points that did not correspond to the metal alloy fcc phase (i.e., where Phase ≠ 1), the data row for that point in the *.ctf file was overwritten with a new Phase identifier and dummy values for the crystallographic information. These dummy values for orientation persist into the final DREAM.3D data file associated with the AM Model Challenge 4 [6], and thus the only valid orientation data in this file are for the fcc phase (Phase = 1). A representative image of the fused *.ctf data is shown in Fig. 6h.
3D Reconstruction of Fused SS Data
The fused *.ctf data were used as input and further processed using DREAM.3D software [21] to create the 3D reconstruction of the HEDM VOI used in the AM Model Challenge. Key tasks of this process included the following: registration of the reference frames of the SS data volume to the far-field HEDM data previously collected from the synchrotron in-situ experiment, segmentation of grains and clean up of unassigned data points, and homogenization of the grain orientation data. The remainder of this section expands on the 3D reconstruction procedure with additional details.
The 3D reconstruction process began by reading the the fused *.ctf data into DREAM.3D as a data volume, stacking the sections from high to low (to account for the SS file labeling scheme where later sections have an increasing number value), with voxel dimensions of 1.0 × 1.0 × 0.97 µm in x, y and z, respectively and the z-axis corresponding to both the SS and the AM build direction. Additionally, the Oxford file format was selected when setting the reference frame, in order to rotate the sample 180° about <010> to bring the sample reference frame in coincidence with the crystallographic reference frame. For the remainder of this workflow, the sample and crystallographic reference frame are identical. The authors note that three sections from the complete SS experiment contained EBSD data of poor quality or significant artifacts during automated data collection. However, two of the flagged sections were outside the subvolume that was ultimately used in the Challenge 4 reconstruction, and thus the authors assumed the error due to this replacement was minimal after the aforementioned subvolume cropping and additional spatial downsampling. Next, the DREAM.3D “Reduce Orientations to Fundamental Zone” filter was used to reduce all the EBSD orientation measurements to the fundamental zone for the fcc crystal structure, to aid in correlating the spatial and orientation reference frame of the SS data to the far-field HEDM data.
The process for optimizing the registration of the SS and HEDM data was somewhat complex and required significant human intervention and data manipulation. A prior study by Lenthe and colleagues has addressed a similar challenge of registering SS to diffraction-contrast tomography (DCT) data [22], although the internal microstructure for the present study is comparatively more complex. An initial challenge for the authors was to ensure that the two data volumes (SS and HEDM) were providing similar representations and reconstructions of the complex, highly twinned grain microstructure. To accomplish this task, the central region of the tensile sample VOI was examined in detail, where the center gold marker denoted a small subvolume where a near-field HEDM (nf-HEDM) reconstruction (collected before in-situ mechanical testing) had been reconstructed from line-focus beam data [3]. In order to bring the SS data into general alignment with the nf-HEDM data, a rotation of – 90° about [001] was applied to one section of EBSD data at the center of the gold marker. Again, this rotation was performed for the purpose of determining the minor orientation differences between the EBSD and HEDM measurements that could not be accounted for by reference frame shifts.
Figure 7 shows a comparison of the nf-HEDM reconstruction and SS EBSD map from the central Au marker region (using Paraview to visualize the Euler angle data from both data sets). Visual examination of Fig. 7 shows that selected grains can be clearly identified in both data sets, although there are noticeable differences in the number and spatial arrangement of annealing twins for the majority of grains, as well as clear differences in the microstructure at the outer edge of the tensile sample. Quantitative analysis of the differences between the data sets was not further explored, and remains a topic of future interest. The degree of orientation misalignment between the data sets was quantified by manual selection of 10 points from 10 different grains that were clearly identifiable in both sets of data, from which an average misorientation value was calculated to be approximately 4.2°.
HEDM and SS characterization data from the tensile sample VOI that correspond to the central gold marker. a nf-HEDM reconstruction before the in-situ mechanical test (for confidence index > 0.5), and b EBSD map from SS data after the in-situ test (IPF coloring, X-direction). One can observe qualitative agreement between the two data sets that facilitated manual selection of grain data points that were readily identified in both data sets, and this information was used to improve the registration between the data volumes as described in the accompanying text
One aspect of the SS experiment that influenced this misalignment was the small tilt of the tensile axis relative to the SS reference frame, which was initially observed during radiographic imaging of the consolidated materialographic mount. The vector representing the tensile axis in the SS data was determined by calculating the vector endpoints from the center of the tensile sample cross-section from the first and last serial section, v_s. The tilt was determined by the cross product of v_s and v_z (the positive z-direction in the SS reference frame), resulting in an axis-angle pair of [− 0.93, 0.37, 0] and 1.80°. Correcting for this slight misalignment reduced the difference between the SS and HEDM data to an average misorientation of approximately 2.6°. The final orientation misalignment was addressed by using an optimization analysis using a custom python script to minimize the integrated point-to-point misorientation between the data sets. The axis-angle pair for this remaining misalignment was determined to be 2.6° about [− 0.93, − 0.027, − 0.36]. The authors note that the SEM stage tilt corresponds to rotation about the x-axis, and recognize that some of the orientation discrepancy could be the result of minor errors in tilt positioning, but this possible cause is unverified at this time. Nevertheless, this final rotation correction was only applied to the SS crystallographic reference frame and not applied to the sample geometry, in order to optimize the orientation registration between SS and HEDM data. Again, this final rotation operation was applied to only the local orientation values of the SS data using the DREAM.3D filter “Rotate Euler Reference Frame.” Collectively, these procedures minimized the average orientation difference between the HEDM and SS data to ~ 0.60°.
Further processing of the 3D reconstruction was performed within DREAM.3D. After applying the various minor rotations described in the previous paragraph to the 3D reconstructed volume, the orientations were again reduced to the fundamental zone. For clarity, we remind the reader that the general reference frame at this point was still the SS reference frame and not the nf- or ff-HEDM reference frame. Those rotations are applied toward the end of the workflow as will be discussed later in this section. The data volume was segmented using the “Segment Features (Misorientation)” filter to define the crystalline regions within the tensile sample using the default 5° misorientation tolerance. For voxels where the Phase value was not equal to 1, the orientations had identical (dummy) values, and therefore were also effectively segmented by this operation. Next, feature sizes were calculated using the “Find Feature Sizes” filter, and any feature that was 7 voxels or smaller was removeds using the “Minimum Size” filter, and those voxels were isotropically absorbed by neighboring features. Minor speckle noise from the surface of the tensile sample (Phase 1) was cleaned by flagging and removing surface features smaller than 2 voxels using the “Find Surface Features” and “Threshold Objects” filters. Segmentation was done again, using the “Segment Features (Misorientation)” filter to identify the new features after the small and surface feature cleanups. Lastly, for each feature, the average orientation was calculated using the “Find Feature Average Orientations” and stored in the FeatureData array.
Final steps in preparing the SS data for the Challenge 4 input included downsampling the data to create voxels that were isotropic with dimensions of 2 µm using the “Change Resolution” filter. The data volume was also cropped using the “Crop Geometry” filter along the sectioning/build direction to ~ 300 µm above and below the center gold marker. Noise corrections were also performed on the gold and platinum Phases to reassign voxels that were incorrectly assigned to one or the other, both by only keeping the large (> 200 voxels) features and by manually identifying incorrectly labeled regions, using the “Initialize Data Array” and “Threshold Objects” filters to remove them. This had no effect on either the tensile sample (Phase 1) or porosity (Phase 2) features and was largely for visual effect. Lastly, additional rotations to the sample and orientations data was performed to transform the reference frame to the convention used for the far-field HEDM data. These rotations were – 90° about [001] to transform the data from the SS reference frame into the nf-HEDM reference frame, followed by a rotation of 120° about [− 111] to transform the data from the nf-HEDM reference frame into the ff-HEDM reference frame. These two rotations were applied to both the sample and orientation data, resulting in the y-direction now being assigned to the SS and build direction. For the challenge data volume, the local orientation values were reduced to the average orientation in each grain, and therefore only the grain average values are provided (which have replaced the original local orientation values). In addition, the initial 6-component strain tensor for each of the selected 28 Challenge 4 grains was also inserted into the grain-level data in the DREAM.3D file. Additional information of the procedure of selecting and matching the Challenge 4 grains from both HEDM to SS data are described in [3]
Figure 8 shows 3D reconstructions using ParaView of the (a) Challenge 4 data volume and (b) the interior 28 grains that were the features-of-interest for the 4th Challenge, where the grain coloring in both images are for the Inverse Pole Figure-Z direction. All of the processing steps that occurred within the DREAM.3D software described above are also recorded in the ‘Pipeline’ group of the associated DREAM.3D file, and can be viewed with any software program that is capable of reading and viewing an HDF5 file. In addition, if the text in the ‘Pipeline’ group is saved as a new *.json file, this file can be read into DREAM.3D as a saved pipeline for further query or use. This pipeline has been checked for compatibility with the following version of DREAM.3D (Version 6.5.128 from Revision cbf99), and the source code for DREAM.3D is publically available on GitHub (https://github.com/BlueQuartzSoftware/DREAM3D). Additional descriptions of the Challenge 4 data package, including both the DREAM.3D data file and the answer key for the Challenge 4 data, can be accessed at the Materials Data Facility (MDF) [6]. The authors note that data retention policy of the MDF is to guarantee the availability of hosted data for at least five years after the ending of the MDF project, and at present this enables public data access until at least the year 2028.
a Visualization of the 3D volume provided as input data for Challenge 4 and b rendering of only the 28 selected challenge grain (IPF coloring, Z-direction). The dimensions of the outlined parallel piped volume are 610 × 702 × 624 µm (with isotropic voxel dimensions of 2 µm), and the sample and crystallographic reference frames are the same
Summary
This data descriptor article documents a serial sectioning (SS) microscopy characterization experiment that was part of the Air Force Research Laboratory Additive Manufacturing (AM) Modeling Series. The article describes the experimental and post-processing workflows to provide a micron-level characterization of the internal microstructure of an IN625 tensile sample using a custom, automated, and multi-modal SS microscopy system at AFRL. The tensile sample had undergone in-situ synchrotron-based High-Energy Diffraction Microscopy (HEDM) characterization to quantify the evolution of local internal strains within individual grains during a mechanical test, which was the focus of Challenge 4 of the AM Modeling Series. Custom post-processing and 3D reconstruction workflows were developed to register and fuse all of the SS data, and a 2 µm voxel resolution reconstruction of the tensile sample microstructure was prepared for the Challenge 4 data package, which can be obtained at the Materials Data Facility.
References
Groeber M (2018) A preview of the U.S. Air Force Research Laboratory additive manufacturing modeling challenge series. JOM 70(4):441–444. https://doi.org/10.1007/s11837-018-2806-3
Schuren JC et al (2015) New opportunities for quantitative tracking of polycrystal responses in three dimensions. Curr Opin Solid State Mater Sci 19(4):235–244. https://doi.org/10.1016/j.cossms.2014.11.003
Menasche DB et al (2021) AFRL additive manufacturing modeling series: challenge 4, in situ mechanical test of an IN625 sample with concurrent high-energy difraction microscopy characterization. Integr Mater Manuf Innov
Blaiszik B, Chard K, Pruyne J, Ananthakrishnan R, Tuecke S, Foster I (2016) The materials data facility: data services to advance materials science research. JOM 68(8):2045–2052. https://doi.org/10.1007/s11837-016-2001-3
Blaiszik B et al (2019) A data ecosystem to support machine learning in materials science. MRS Commun 9(4):1125–1133. https://doi.org/10.1557/mrc.2019.118
Shade PA et al (2019) AFRL AM modeling challenge series: challenge 4 data package. Mater Data Facil. https://doi.org/10.18126/K5R2-32IU
Menasche DB, Shade PA, Suter RM (2020) Accuracy and precision of near-field high-energy diffraction microscopy forward-model-based microstructure reconstructions. J Appl Crystallogr 53(1):107–116. https://doi.org/10.1107/S1600576719016005
Shade PA et al (2016) Fiducial marker application method for position alignment of in situ multimodal X-ray experiments and reconstructions. J Appl Cryst. 49:700–704. https://doi.org/10.1107/S1600576716001989
Madison JD, Underwood OD, Poulter GA, Huffman EM (2017) Acquisition of real-time operation analytics for an automated serial sectioning system. Integr Mater Manuf Innov 6(2):135–146. https://doi.org/10.1007/s40192-017-0091-6
Uchic M, et al (2012) An automated multi-modal serial sectioning system for characterization of grain-scale microstructures in engineering materials. In: Proceedings of the 1st international conference on 3D materials science, Springer, Cham, pp 195–202. https://doi.org/10.1007/978-3-319-48762-5_30
Boyce BL, Uchic MD (2019) Progress toward autonomous experimental systems for alloy development. MRS Bull 44(4):273–280. https://doi.org/10.1557/mrs.2019.75
Kral MV, Spanos G (1999) Three-dimensional analysis of proeutectoid cementite precipitates. Acta Mater 47(2):711–724. https://doi.org/10.1016/S1359-6454(98)00321-8
Russ JC, Neal FB (2018) The image processing handbook, 7th edn. CRC Press, London
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. Int J Comput Vision 60(2):91–110. https://doi.org/10.1023/B:VISI.0000029664.99615.94
Jackson MA, Pascal E, De Graef M (2019) Dictionary indexing of electron back-scatter diffraction patterns: a hands-on tutorial. Integr Mater Manuf Innov 8(2):226–246. https://doi.org/10.1007/s40192-019-00137-4
Nolze G (2007) Image distortions in SEM and their influences on EBSD measurements. Ultramicroscopy 107(2):172–183. https://doi.org/10.1016/j.ultramic.2006.07.003
Fiji: an open-source platform for biological-image analysis | Nature Methods. https://www.nature.com/articles/nmeth.2019. Accessed January 28, 2021
Preibisch S, Saalfeld S, Tomancak P (2009) Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25(11):1463–1465. https://doi.org/10.1093/bioinformatics/btp184
Yaniv Z, Lowekamp BC, Johnson HJ, Beare R (2018) SimpleITK image-analysis notebooks: a collaborative environment for education and reproducible research. J Digit Imaging 31(3):290–303. https://doi.org/10.1007/s10278-017-0037-8
Gulsoy EB, Simmons JP, De Graef M (2009) Application of joint histogram and mutual information to registration and data fusion problems in serial sectioning microstructure studies. Scr Mater 60(6):381–384. https://doi.org/10.1016/j.scriptamat.2008.11.004
Groeber MA, Jackson MA (2014) DREAM.3D: a digital representation environment for the analysis of microstructure in 3D. Integr Mater 3(1):56–72. https://doi.org/10.1186/2193-9772-3-5
Lenthe WC, Echlin MP, Trenkle A, Syha M, Gumbsch P, Pollock TM (2015) Quantitative voxel-to-voxel comparison of TriBeam and DCT strontium titanate three-dimensional data sets. J Appl Crystallogr 48(4):1034–1046. https://doi.org/10.1107/S1600576715009231
Acknowledgements
All of the authors acknowledge support from the Air Force Research Laboratory. MC and JMS acknowledge support from the Air Force Research Laboratory through contract FA8650-19-F-5205, and DM acknowledges support from the Air Force Research Laboratory through contract FA8650-20-F-5212. The authors acknowledge technical contributions on providing the AM sample and discussion with Edwin Schwalbach and Marie Cox of AFRL, and Michael Groeber of The Ohio State University (when previously employed at AFRL). The authors gratefully acknowledge the technical contributions of Andy Newman at NASA-Langley for Plasma-Focused Ion Beam microscopy, Sirina Safriet of UDRI for FIB microscopy, Daniel Sparkman of AFRL for radiographic image collection, Marc De Graef of Carnegie Mellon University, and Michael Jackson from BlueQuartz Software for dictionary reindexing of the EBSD data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chapman, M.G., Shah, M.N., Donegan, S.P. et al. AFRL Additive Manufacturing Modeling Series: Challenge 4, 3D Reconstruction of an IN625 High-Energy Diffraction Microscopy Sample Using Multi-modal Serial Sectioning. Integr Mater Manuf Innov 10, 129–141 (2021). https://doi.org/10.1007/s40192-021-00212-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40192-021-00212-9