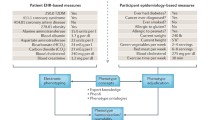
Abstract
With the large volume of clinical and epidemiological data being collected, increasingly linked to extensive genotypic data, coupled with expanding high-performance computational resources, there are considerable opportunities for comprehensively exploring the networks of connections that exist between the phenome and the genome. These networks can be identified through Phenome-Wide Association Studies (PheWAS) where the association between a collection of genetic variants, or in some cases a particular clinical lab variable, and a wide and diverse range of phenotypes, diagnoses, traits, and/or outcomes are evaluated. This is a departure from the more familiar genome-wide association study approach, which has been used to identify single nucleotide polymorphisms associated with one outcome or a very limited phenotypic domain. In addition to highlighting novel connections between multiple phenotypes and elucidating more of the phenotype-genotype landscape, PheWAS can generate new hypotheses for further exploration, and can also be used to narrow the search space for research using comprehensive data collections. The complex results of PheWAS also have the potential for uncovering new mechanistic insights. We review here how the PheWAS approach has been used with data from epidemiological studies, clinical trials, and de-identified electronic health record data. We also review methodologies for the analyses underlying PheWAS, and emerging methods developed for evaluating the comprehensive results of PheWAS including genotype–phenotype networks. This review also highlights PheWAS as an important tool for identifying new biomarkers, elucidating the genetic architecture of complex traits, and uncovering pleiotropy. There are many directions and new methodologies for the future of PheWAS analyses, from the phenotypic data to the genetic data, and herein we also discuss some of these important future PheWAS developments.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as follows: • Of importance •• Of major importance
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106:9362–7.
Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254.
Knight JC. Genomic modulators of the immune response. Trends Genet. 2013;29:74–83.
Kraja AT, Chasman DI, North KE, Reiner AP, Yanek LR, Kilpeläinen TO, et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014;112:317–8.
•• Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. First PheWAS, used electronic health record data.
Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42.
Hebbring SJ, Schrodi SJ, Ye Z, Zhou Z, Page D, Brilliant MH. A PheWAS approach in studying HLA-DRB1*1501. Genes Immun. 2013;14:187–91.
Liao KP, Kurreeman F, Li G, Duclos G, Murphy S, Guzman R, et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum. 2013;65:571–81.
Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109.
Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10.
Verma A, Kuivaniemi H, Tromp G, Carey DJ, Gerhard GS, Crowe JE, et al. Exploring the relationship between immune system related genetic variants and complex traits and disease through a Phenome-Wide Association Study (PheWAS) (under review).
Ye Z, Mayer J, Ivacic L, Zhou Z, He M, Schrodi SJ, et al. Phenome-wide association studies (PheWASs) for functional variants. Eur J Hum Genet. 2014;23:523–9.
•• Pendergrass SA, Brown-Gentry K, Dudek S, Frase A, Torstenson ES, Goodloe R, et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 2013;9:e1003087. First PheWAS using epidemiological study data.
Hall MA, Verma A, Brown-Gentry K, Goodloe RJ, Boston J, Wilson S, et al. Detection of pleiotropy through a phenome-wide association study (PheWAS) of epidemiologic data as part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study. PLoS Genet. 2014;10:e1004678.
•• Moore CB, Verma A, Pendergrass S, Verma SS, Johnson DH, Daar ES, et al. Phenome-wide Associations Study (PheWAS) relating pre-treatment laboratory parameters with human genetic variants in AIDS clinical trials group protocols. Open Forum Infect Dis. 2014. doi:10.1093/ofid/ofu113. First PheWAS using clinical trials data.
•• Mitchell SL, Hall JB, Goodloe RJ, Boston J, Farber-Eger E, Pendergrass SA, et al. Investigating the relationship between mitochondrial genetic variation and cardiovascular-related traits to develop a framework for mitochondrial phenome-wide association studies. BioData Mining. 2014;7:6. First PheWAS using mitochondrial genetic variation.
Neuraz A, Chouchana L, Malamut G, Le Beller C, Roche D, Beaune P, et al. Phenome-wide association studies on a quantitative trait: application to TPMT enzyme activity and thiopurine therapy in pharmacogenomics. PLoS Comput Biol. 2013;9:e1003405.
• Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. First Environment-Wide Association Study (EWAS).
Hall MA, Dudek SM, Goodloe R, Crawford DC, Pendergrass SA, Peissig P, et al. Environment-wide association study (EWAS) for type 2 diabetes in the Marshfield Personalized Medicine Research Project Biobank. Pac Symp Biocomput. 2014;19:200–11.
• Davis MA, Gilbert-Diamond D, Karagas MR, Li Z, Moore JH, Williams SM, et al. A Dietary-Wide Association Study (DWAS) of environmental metal exposure in US children and adults. PLoS One. 2014;9:e104768. First Dietary-Wide Association Study.
Shuman HA, Silhavy TJ. The art and design of genetic screens: Escherichia coli. Nat Rev Genet. 2003;4:419–31.
Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94.
Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24:103–6.
Bilder RM, Sabb FW, Cannon TD, London ED, Jentsch JD, Parker DS, et al. Phenomics: the systematic study of phenotypes on a genome-wide scale. Neuroscience. 2009;164:30–42.
• Lanktree MB, Hassell RG, Lahiry P, Hegele RA. Phenomics: expanding the role of clinical evaluation in genomic studies. J Investig Med. 2010;58:700–6. Review/idea paper about leveraging phenomics for improved understanding of disease.
• Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–66. Detailed review of considerations for phenomics.
Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabasi A-L. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–90.
Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA. 2007;104:11694–9.
• Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. Review of exploring networks and interactions for understanding disease.
Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013;29:66–73.
•• Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–95. Excellent review of the importance of pleiotropy and cross-phenotype associations.
•• Jones R, Pembrey M, Golding J, Herrick D. The search for genenotype/phenotype associations and the phenome scan. Paediatr Perinat Epidemiol. 2005;19:264–75. Idea paper introducing fundamental ideas behind PheWAS focused on networks in the context of dense phenotypic and genotypic information.
•• Ghebranious N, McCarty CA, Wilke RA. Clinical phenome scanning. Personal Med. 2007;4:175–82. Idea paper introducing fundamental ideas behind PheWAS, particularly with comprehensive electronic health record data.
Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–9.
Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–71.
Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG, et al. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5:184.
Verma A, Verma SS, Pendergrass SA, Crawford DC, Crosslin DR, Kuivaniemi H, et al. Phenome-Wide Association Study (PheWAS) identifies clinical associations and pleiotropy for functional variants (in preparation).
• Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011;35:410–22. Idea paper describing PheWAS in epidemiological study based data sets.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
McCarty CA, Huggins W, Aiello AE, Bilder RM, Hariri A, Jernigan TL, et al. PhenX RISING: real world implementation and sharing of PhenX measures. BMC Med Genomics. 2014;7:16.
Hsu C-N, Kuo C-J, Cai C, Pendergrass SA, Ritchie MD, Ambite JL. Learning phenotype mapping for integrating large genetic data. In: Proceedings of BioNLP 2011 Workshop. Portland: Association for Computational Linguistics; 2011. pp. 19–27.
Stephens M. A Unified Framework for Association Analysis with Multiple Related Phenotypes. PLoS One. 2013;8:e65245.
Kim S, Xing EP. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS Genet. 2009;5:e1000587.
Tyler AL, Crawford DC, Pendergrass SA. Detecting and characterizing pleiotropy: new methods for uncovering the connection between the complexity of genomic architecture and multiple phenotypes- session introduction. Pac Symp Biocomput. 2014;19:183–7.
Tyler AL, Lu W, Hendrick JJ, Philip VM, Carter GW. CAPE: an R package for combined analysis of pleiotropy and epistasis. PLoS Comput Biol. 2013;9:e1003270.
Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media. 2009.
Darabos C, White MJ, Graham BE, Leung DN, Williams SM, Moore JH. The multiscale backbone of the human phenotype network based on biological pathways. BioData Min. 2014;7:1.
Darabos C, Harmon SH, Moore JH. Using the bipartite human phenotype network to reveal pleiotropy and epistasis beyond the gene. Pac Symp Biocomput. 2014;19:188–99.
• Williams SM, Haines JL. Correcting away the hidden heritability. Ann Human Genet. 2011;75:348–50. Important considerations for GWAS multiple testing also important for PheWAS.
Sobota RS, Shriner D, Kodaman N, Goodloe R, Zheng W, Gao YT, et al. Addressing population-specific multiple testing burdens in genetic association studies. Ann Human Genet. 2015;79(2):136–47.
Disclosures
SA Pendergrass declares no conflicts of interest. MD Ritchie has received research grants from the NIH and Geisinger Health Systems.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Genomics.
Rights and permissions
About this article
Cite this article
Pendergrass, S.A., Ritchie, M.D. Phenome-Wide Association Studies: Leveraging Comprehensive Phenotypic and Genotypic Data for Discovery. Curr Genet Med Rep 3, 92–100 (2015). https://doi.org/10.1007/s40142-015-0067-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-015-0067-9