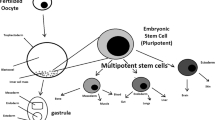
Abstract
Organoids are three-dimensional culture systems that resemble their organ of origin, are genetically stable, and can phenocopy diseases. They enable modeling of various cancer entities such as gastric or colorectal cancer, in addition to other gastrointestinal tract diseases such as inflammatory bowel disease. Genetic engineering tools like CRISPR/Cas9 allow their manipulation to repair mutations or unravel gene functions. Individual patient-derived organoids allow to test therapies in vitro before their in vivo application, bringing personalized medicine to a next level. Organoid biobanks can be used to conduct drug screenings and validate biomarkers. Interactions with microbiota can be investigated in realistic in vitro models. Transplantability of genetically engineered organoids opens up new avenues in the tissue engineering research field. Organoid cultures thus represent a versatile system to model diseases and test therapeutic interventions.

Similar content being viewed by others
References
Recently published papers of particular interest have been highlighted as: • Of importance
• Lancaster MA, Knoblich JA (2014) Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345(6194):1247125. Excellent review about differentiation of PSCs and AdSCs into various organoid culture systems
Huch M, Koo BK (2015) Modeling mouse and human development using organoid cultures. Development 142(18):3113–3125
Lancaster MA, Renner M, Martin CA et al (2013) Cerebral organoids model human brain development and microcephaly. Nature 501(7467):373–379
Muguruma K, Nishiyama A, Kawakami H et al (2015) Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep 10(4):537–550
Koehler KR, Mikosz AM, Molosh AI et al (2013) Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500(7461):217–221
Lee GY, Kenny PA, Lee EH et al (2007) Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4(4):359–365
Dontu G, Abdallah WM, Foley JM et al (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17(10):1253–1270
Suga H, Kadoshima T, Minaguchi M et al (2011) Self-formation of functional adenohypophysis in three-dimensional culture. Nature 480(7375):57–62
Eiraku M, Takata N, Ishibashi H et al (2011) Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472(7341):51–56
Nakano T, Ando S, Takata N et al (2012) Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10(6):771–785
Kuwahara A, Ozone C, Nakano T et al (2015) Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat Commun 6:6286
Hisha H, Tanaka T, Kanno S et al (2013) Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci Rep 3:3224
Stevens KR, Pabon L, Muskheli V et al (2009) Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue Eng Part A 15(6):1211–1222
Takasato M, Er PX, Becroft M et al (2014) Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol 16(1):118–126
Batchelder CA, Martinez ML, Duru N et al (2015) Three dimensional culture of human renal cell carcinoma organoids. PLoS One 10(8):e0136758
Kwong J, Chan FL, Wong KK et al (2009) Inflammatory cytokine tumor necrosis factor alpha confers precancerous phenotype in an organoid model of normal human ovarian surface epithelial cells. Neoplasia 11(6):529–541
van der Schaft DWJ, van Spreeuwel ACC, Boonen KJM et al (2013) Engineering skeletal muscle tissues from murine myoblast progenitor cells and application of electrical stimulation. JoVE 73:4267
Spence JR, Mayhew CN, Rankin SA et al (2011) Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470(7332):105–109
Sato T, Vries RG, Snippert HJ et al (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244):262–265
Ootani A, Li X, Sangiorgi E et al (2009) Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med 15(6):701–706
Sato T, Stange DE, Ferrante M et al (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141(5):1762–1772
• Li X, Nadauld L, Ootani A et al (2014) Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med 20(7):769–777. Special air-liquid interface methodology for the culturing of complex gastric, pancreatic and intestinal organoids without growth factors and various mutation studies for validation of tumorigenicity in vitro and in vivo
• van de Wetering M, Francies HE, Francis JM et al (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161(4):933–945. Establishment of an extensive “Living biobank” with CRC patient-derived organoids applied for DNA sequencing, transcriptome analysis and personalized drug screening
Takebe T, Sekine K, Enomura M et al (2013) Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499(7459):481–484
Huch M, Dorrell C, Boj SF et al (2013) In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494(7436):247–250
Huch M, Gehart H, van Boxtel R et al (2015) Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160(1–2):299–312
Dye BR, Hill DR, Ferguson MA et al (2015) In vitro generation of human pluripotent stem cell derived lung organoids. Elife. doi:10.7554/eLife.05098
Lee JH, Bhang DH, Beede A et al (2014) Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156(3):440–455
Greggio C, De Franceschi F, Figueiredo-Larsen M et al (2013) Artificial three-dimensional niches deconstruct pancreas development in vitro. Development 140(21):4452–4462
Huch M, Bonfanti P, Boj SF et al (2013) Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32(20):2708–2721
Boj SF, Hwang CI, Baker LA et al (2015) Organoid models of human and mouse ductal pancreatic cancer. Cell 160(1–2):324–338
Xin L, Lukacs RU, Lawson DA et al (2007) Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells 25(11):2760–2769
Karthaus WR, Iaquinta PJ, Drost J et al (2014) Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159(1):163–175
Gao D, Vela I, Sboner A et al (2014) Organoid cultures derived from patients with advanced prostate cancer. Cell 159(1):176–187
McCracken KW, Cata EM, Crawford CM et al (2014) Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516(7531):400–404
Barker N, Huch M, Kujala P et al (2010) Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6(1):25–36
Stange DE, Koo BK, Huch M et al (2013) Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155(2):357–368
Bartfeld S, Bayram T, van de Wetering M et al (2015) In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148(1):126–136, e126
Antonica F, Kasprzyk DF, Opitz R et al (2012) Generation of functional thyroid from embryonic stem cells. Nature 491(7422):66–71
Koo BK, Clevers H (2014) Stem cells marked by the R-spondin receptor Lgr5. Gastroenterology 147(2):289–302
Andersson-Rolf A, Fink J, Mustata RC et al (2014) A video protocol of retroviral infection in primary intestinal organoid culture. J Vis Exp 90:e51765
Drost J, van Jaarsveld RH, Ponsioen B et al (2015) Sequential cancer mutations in cultured human intestinal stem cells. Nature 521(7550):43–47
Koo BK, Stange DE, Sato T et al (2012) Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods 9(1):81–83
• Matano M, Date S, Shimokawa M et al (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21(3):256–262. Modeling of the most common CRC mutations via CRISPR/Cas9 genome editing, establishment of a media-based selection method and validation of the tumorigenicity in vitro and in vivo
Schwank G, Andersson-Rolf A, Koo BK et al (2013) Generation of BAC transgenic epithelial organoids. PLoS One 8(10):e76871
• Schwank G, Koo BK, Sasselli V et al (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13(6):653–658. Repair of a disease-causing mutation by homologous recombination with CRISPR/Cas9
• Yui S, Nakamura T, Sato T et al (2012) Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med 18(4):618–623. First study showing the transplantability of organoid cultures for the repair of damaged tissue
Torre LA, Bray F, Siegel RL et al (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108
Cancer Genome Atlas N (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407):330–337
Sachs N, Clevers H (2014) Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev 24:68–73
Barker N, van Es JH, Kuipers J et al (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449(7165):1003–1007
Munoz J, Stange DE, Schepers AG et al (2012) The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J 31(14):3079–3091
Radtke F, Clevers H (2005) Self-renewal and cancer of the gut: two sides of a coin. Science 307(5717):1904–1909
Chen EC, Karl TA, Kalisky T et al (2015) KIT signaling promotes growth of colon xenograft tumors in mice and is up-regulated in a subset of human colon cancers. Gastroenterology 149(3):705–717, e702
Calon A, Lonardo E, Berenguer-Llergo A et al (2015) Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47(4):320–329
Wiener Z, Band AM, Kallio P et al (2014) Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-beta. Proc Natl Acad Sci USA 111(21):E2229–E2236
Yamauchi M, Otsuka K, Kondo H et al (2014) A novel in vitro survival assay of small intestinal stem cells after exposure to ionizing radiation. J Radiat Res 55(2):381–390
Tomono A, Yamashita K, Kanemitsu K et al (2015) Prognostic significance of pathological response to preoperative chemoradiotherapy in patients with locally advanced rectal cancer. Int J Clin Oncol. doi:10.1007/s10147-015-0900-x
Vassilev LT, Vu BT, Graves B et al (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303(5659):844–848
Grivennikov SI, Greten FR, Karin M (2010) Immunity, inflammation, and cancer. Cell 140(6):883–899
Oshima H, Nakayama M, Han TS et al (2015) Suppressing TGFbeta signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res 75(4):766–776
Koo BK, Spit M, Jordens I et al (2012) Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488(7413):665–669
Wong VW, Stange DE, Page ME et al (2012) Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14(4):401–408
van der Flier LG, van Gijn ME, Hatzis P et al (2009) Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136(5):903–912
Schuijers J, van der Flier LG, van Es J et al (2014) Robust cre-mediated recombination in small intestinal stem cells utilizing the olfm4 locus. Stem Cell Reports 3(2):234–241
Huch M (2015) Building stomach in a dish. Nat Cell Biol 17(8):966–967
Noguchi TA, Ninomiya N, Sekine M et al (2015) Generation of stomach tissue from mouse embryonic stem cells. Nat Cell Biol 17(8):984–993
Nadauld LD, Garcia S, Natsoulis G et al (2014) Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol 15(8):428
Salama NR, Hartung ML, Muller A (2013) Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol 11(6):385–399
Schlaermann P, Toelle B, Berger H et al (2014) A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 65(2):202–213
Schumacher MA, Feng R, Aihara E et al (2015) Helicobacter pylori-induced Sonic Hedgehog expression is regulated by NFkappaB pathway activation: the use of a novel in vitro model to study epithelial response to infection. Helicobacter 20(1):19–28
Sigal M, Rothenberg ME, Logan CY et al (2015) Helicobacter pylori activates and expands Lgr5(+) stem cells through direct colonization of the gastric glands. Gastroenterology 148(7):1392–1404, e1321
Marchesi JR, Adams DH, Fava F et al (2015) The gut microbiota and host health: a new clinical frontier. Gut 65(2):330–339
Lukovac S, Belzer C, Pellis L et al (2014) Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. doi:10.1128/mBio.01438-14
Finkbeiner SR, Zeng XL, Utama B et al (2012) Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 3(4):e00159–00112
Yin Y, Bijvelds M, Dang W et al (2015) Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res 65(2):330–339
Zhang YG, Wu S, Xia Y et al (2014) Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep. doi:10.14814/phy2.12147
Engevik MA, Aihara E, Montrose MH et al (2013) Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol 305(10):G697–711
Engevik MA, Engevik KA, Yacyshyn MB et al (2015) Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol 308(6):G497–509
Engevik MA, Yacyshyn MB, Engevik KA et al (2015) Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 308(6):G510–524
Leslie JL, Huang S, Opp JS et al (2015) Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun 83(1):138–145
Wilson SS, Tocchi A, Holly MK et al (2015) A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol 8(2):352–361
Viswanathan VK (2014) Muramyl dipeptide: not just another brick in the wall. Gut Microbes 5(3):275–276
Nigro G, Rossi R, Commere PH et al (2014) The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 15(6):792–798
Pham TA, Clare S, Goulding D et al (2014) Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 16(4):504–516
Li Y, Teo WL, Low MJ et al (2014) Constitutive TLR4 signalling in intestinal epithelium reduces tumor load by increasing apoptosis in APC(Min/+) mice. Oncogene 33(3):369–377
Rodansky ES, Johnson LA, Huang S et al (2015) Intestinal organoids: a model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp Mol Pathol 98(3):346–351
Schumacher MA, Aihara E, Feng R et al (2015) The use of murine-derived fundic organoids in studies of gastric physiology. J Physiol 593(8):1809–1827
Mokry M, Middendorp S, Wiegerinck CL et al (2014) Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology 146(4):1040–1047
Dammann K, Khare V, Lang M et al (2015) PAK1 modulates a PPARgamma/NF-kappaB cascade in intestinal inflammation. Biochim Biophys Acta 1853(10 Pt A):2349–2360
Fordham RP, Yui S, Hannan NR et al (2013) Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 13(6):734–744
Fukuda M, Mizutani T, Mochizuki W et al (2014) Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev 28(16):1752–1757
Bigorgne AE, Farin HF, Lemoine R et al (2014) TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J Clin Invest 124(1):328–337
Zomer-van Ommen DD, Vijftigschild LA, Kruisselbrink E et al (2015) Limited premature termination codon suppression by read-through agents in cystic fibrosis intestinal organoids. J Cyst Fibros
Trzcinska-Daneluti AM, Chen A, Nguyen L et al (2015) RNA interference screen to identify kinases that suppress rescue of DeltaF508-CFTR. Mol Cell Proteomics 14(6):1569–1583
Okiyoneda T, Veit G, Dekkers JF et al (2013) Mechanism-based corrector combination restores DeltaF508-CFTR folding and function. Nat Chem Biol 9(7):444–454
Dekkers JF, van der Ent CK, Beekman JM (2013) Novel opportunities for CFTR-targeting drug development using organoids. Rare Dis 1:e27112
Dekkers JF, Wiegerinck CL, de Jonge HR et al (2013) A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 19(7):939–945
Mou H, Brazauskas K, Rajagopal J (2015) Personalized medicine for cystic fibrosis: establishing human model systems. Pediatr Pulmonol 50(Suppl 40):S14–23
Ikpa PT, Bijvelds MJ, de Jonge HR (2014) Cystic fibrosis: toward personalized therapies. Int J Biochem Cell Biol 52:192–200
Brugmann SA, Wells JM (2013) Building additional complexity to in vitro-derived intestinal tissues. Stem Cell Res Ther 4(Suppl 1):S1
Grant CN, Grikscheit TC (2013) Tissue engineering: a promising therapeutic approach to necrotizing enterocolitis. Semin Pediatr Surg 22(2):112–116
Belchior GG, Sogayar MC, Grikscheit TC (2014) Stem cells and biopharmaceuticals: vital roles in the growth of tissue-engineered small intestine. Semin Pediatr Surg 23(3):141–149
Behjati S, Huch M, van Boxtel R et al (2014) Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 513(7518):422–425
Acknowledgments
The Stange lab is funded by the European Research Council (#1570398.99), Deutsche Krebshilfe (#111350), Wilhelm-Sander-Stiftung (#2014.104.1), and H.W. & J. Hector Stiftung (M 65.2). Figure 1 was designed with the help of http://www.freepik.com.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Organoid Cultures.
Rights and permissions
About this article
Cite this article
Werner, K., Weitz, J. & Stange, D.E. Organoids as Model Systems for Gastrointestinal Diseases: Tissue Engineering Meets Genetic Engineering. Curr Pathobiol Rep 4, 1–9 (2016). https://doi.org/10.1007/s40139-016-0100-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40139-016-0100-z