Abstract
Introduction
Pakistan has one of the highest burdens of pneumococcal diseases in the world, but unfortunately studies in this demanding research area are limited in the region. Pneumococcal surface protein A (PspA) is the next generation pneumococcal vaccine candidate as the protein locates on the Streptococcus pneumoniae surface. Its gene, pspA, might be encoded by all pneumococci, and the protein has proven immunogenicity. The molecular characterization of PspA, pneumococcal serotype distribution and antibiotic susceptibility are important for regional diversity studies.
Methods
In this study, we examined 38 pneumococcal isolates from pneumococcal diseased (pneumonia/meningitis) patients blood or cerebrospinal fluid. There were no specific inclusion or exclusion criteria, but all the individuals [ages 1 month to 12 years (male/female)] had undergone no antibiotic treatment in at least the past 3 months and had no vaccination history. We investigated the serotype distribution, antibiotic susceptibility, prevalence of the PspA family and its active domain's fusion, expression and antigenicity.
Results
Our finding shows that serotype 19F is the most prevalent (23.6%) followed by 18B (15.78%) (non-vaccine type) in all isolated pneumococcal strains. All strains were susceptible to chloramphenicol and linezolid, while 80% were resistant to gentamycin. Genotyping revealed that ~ 80% (N = 31/38) of pneumococcal strains produce PspA belonging to family 2 and clade 3. We further selected three active domains of PspA (family 2 and clade 3) by in silico analysis, merged together into a fusion gene for expression study, and its antigenicity was analyzed by Western blotting.
Conclusion
Serotypes 19F and 18B (non-vaccine type) are the most prevalent in the Pakistani pneumococcal isolates. The PspA family 2 proteins produced by Pakistani pneumococcal isolates have high sequence homologies with each other and differ from those produced by strains isolated in the rest of the world. The PspA fusion peptide had a proven antigenic response in western blotting, with no considerable correlation among pneumococcal serotypes, antibiotic susceptibility and PspA family/clade distribution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptococcus pneumoniae (pneumococcus) causes pneumonia, otitis media and other invasive diseases such as bacteremia and meningitis, collectively called pneumococcal diseases (PD). Pneumococcus plays a major role in the morbidity and mortality of children as well as the elderly worldwide [1]. It is estimated that ~ 14.5 million cases of PD occur each year, causing the deaths of ~ 1 million children < 5 years of age [2]. The increasing antibiotic resistance of S. pneumoniae worldwide is another concern for the treatment of PD [3]. There are more than 90 known pneumococcal serotypes, but most PDs are produced by a few types [4]. Vaccines against these serotypes, which generate immunity against the capsular polysaccharide, have reduced the burden of pneumococcal diseases in some countries [5]. Pneumococcal polysaccharide vaccines (PSVs) protect against numerous serotypes. However, PSV does not generate isotype switching and memory B cell induction, as it is a T-cell-independent immunogen, thus leading to temporary protection, and it is only recommended for the elderly population [6]. To solve this problem, pneumococcal conjugate vaccines (PCVs) were developed, including a carrier-peptide, diphtheria toxoid (CRM-197), which mediates the T-cell response. PCVs initiate memory B cell production, resulting in robust immunity in infants [7]. PCVs cause an active immune response and protect against the most prevalent serotypes, but new approaches are needed because of the phenomenon of serotype replacement post-vaccination, cost and production complications [8, 9]. Therefore, the development of more effective vaccines against pneumococcus with low production costs but having a broader spectrum is warranted. The key is to develop a vaccine that protects against all known pneumococcal serotypes and new strains as they emerge. Some research groups have recently demonstrated that peptide-based vaccines might be useful against PD [9, 10]. Pneumococcal peptide-based vaccines utilize the most immunogenic pneumococcal protein antigens, such as pneumococcal surface protein A (PspA), pneumococcal surface protein C (PspC), pneumolysin (Ply) and pneumococcal surface adhesin A (PsaA), and many others have shown immunogenicity and protection against lethal challenge with pneumococcus in animal models [8].
The pneumococcal surface protein A (PspA) is a surface-exposed protein and one of the candidates for inclusion in the next generation of pneumococcal vaccines as the protein locates on the surface of all pneumococci and is immunogenic. For example, PspA interferes with the fixation of complement C3b and also binds human lactoferrin, which interferes with its protective role [11, 12]. All studied pneumococcal strains carry the gene pspA, which encodes for the protein PspA [13]. When the mature PspA protein was injected in an animal model, anti-PspA antibodies were detected [14]. Molecular studies of the PspA protein have been classically carried out using serotype 2, strain Rx1. The protein (PspA) has five domains, including (1) an N-terminus signal peptide, (2) alpha-helical domain, (3) proline-rich region, (4) choline-binding domain consisting of 20 amino acid repeats and (5) C-terminus tail. PspA utilizes its choline-binding domain to anchor onto the pneumococcal cell surface via lipoteichoic acids and its alpha-helical domain to bind to the surface of host cells via a still unknown receptor. Based on the sequence (clade-defining region) of the alpha-helical region, PspA can be classified into three families, from 1 to 3, and further subdivided into six clades (clades 1–6) [15].
Pakistan, a country located in South Asia, has the third most pneumococcal diseases in the world, especially ‘pneumococcal pneumonia,’ which makes it an ideal location to begin exploring pneumococcal serotypes and antibiotic susceptibility [16]. In this pilot study, we sought to characterize the prevalence of PspA from Pakistani pneumococcal isolates and identify the most prevalent family and clade.
Methods
Compliance with Ethics Guidelines
The current study (both the study and collection of specimens) was approved by the Advanced Studies and Research Board of the University of the Punjab, Lahore, Pakistan (D/4439-ACAD), and sample collection from patients was approved by the Institutional Review Board (IRB)/Ethics Committee of the Children’s Hospital and the Institute of Child Health, Lahore, Pakistan (CH.AD251). Specimens were collected (according to recommended and approved procedures) from those patients or children whose parents or guardians agreed to participate in this study; it was specified that biologic material would only be utilized for research purposes. It was further clarified that all procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Streptococcus Pneumoniae Isolation and Identification of Serotypes
In this study, we examined suspected pneumococcal disease (pneumonia and meningitis) patient's blood or cerebrospinal fluid (CSF) [17], and N = 38 specimens were confirmed positive for pneumococci. All the individuals (aged 1 month to 12 years, male/female, from 2014 to 2015) were newly admitted to the hospital and had undergone no antibiotic treatment in at least the last 3 months and had no vaccination history. There were no specific inclusion or exclusion criteria.
Streptococcus pneumoniae isolates were identified with standard methods including sensitivity to optochin, bile salt solubility, catalase negativity, alpha-hemolysis and the agglutination method using the BBL™ Pneumoslide™ Test (catalog no. 240840) [18, 19]. The isolates were further confirmed as S. pneumoniae using a PCR procedure that targeted the lytA gene [20]. Pneumococcal strain serotyping was done by the quellung reaction using the SSI® Pneumotest kit (Statens Serum Institut, Denmark) [21].
Antibiotic Susceptibility Testing
Antibiotic susceptibility tests were carried out using antimicrobial disks (Oxoid™, UK) following the EUCAST recommendations [22]. Antibiotics were selected according to common prescriptions in Pakistani hospitals and included: ampicillin (25 μg), amikacin (30 μg), amoxycillin (25 μg), co-amoxiclav (amoxicillin/clavulanic acid) (30 μg), cefotaxime (30 μg), ceftriaxone (30 μg), cefuroxime (30 μg), cephalexin (30 μg), chloramphenicol (30 μg), fusidic acid (10 μg), gentamycin (30 μg), linezolid (30 μg), teicoplanin (30 μg) and vancomycin (30 μg).
DNA Extraction and Amplification of the pspA Gene
Streptococcus pneumoniae genomic DNA was extracted from an overnight culture grown on Todd-Hewitt broth with 0.5% yeast extract (Oxoid™, UK) incubated at 37 °C with a 5% CO2 atmosphere using a DNA extraction kit (Invitrogen™, USA) following the manufacturer’s instructions. The pspA gene was identified and amplified by PCR in a 25-μl reaction volume containing 2.5 mM MgCl2, 1X Taq DNA polymerase buffer, 2 mM (each) deoxynucleoside triphosphates (dNTPs), 1 μM of each primer (SKH2 and LSM12) (Table 1), 2.5 U of Taq DNA polymerase (Invitrogen™, USA) and 25–30 ng/μl pneumococcal genomic DNA template. To specifically amplify the pspA gene belonging to the different families, the following pairs of primers were utilized: LSM12 and SKH63 (family 1), LSM12 and SKH52 (family 2) and SKH41 and SKH42 (family 3) (Table 1) [15]. The following PCR conditions were used: an initial denaturing step at 95 °C for 3 min, 30 cycles of 95 °C for 1 min, 62 °C for 1 min and 72 °C for 2 min, followed by a final extension at 72 °C for 10 min. All PCR products obtained from pneumococcal strains encoding family 2 were purified and sequenced using GenScript (GenScript USA, Inc.) as well as the core facility of the Centre of Applied Molecular Biology, Lahore, Pakistan using PspA gene-specific family 2 forward and reverse primers. DNA sequences were assembled using NCBI BLAST online. The clade distributions were found by the clade-defining region (CDR) occurring in the PspA protein.
Evolutionary and Phylogenetic Analysis
The PspA protein and its DNA sequences were observed using the MEGA-6 and ClustalW2 tools for evolutionary and phylogenetic analysis with each other and with the online NCBI databank PspA depository [23, 24]. The PspA sequencing data were analyzed according to known geographical regions available on online NCBI database.
Three-dimensional Structure of PspA Family 2, Clade 3
The hypothetical 3D structure of PspA family 2, clade 3, was required to identify peptides that could have been exposed on the surface; we therefore predicted the structure using the commercially available DNAstar™ software and by protein homology modeling [25].
In-silico Determination of Immunogenic Sites
The physicochemical properties of the amino acids are considered key to calculating antigenic sites and the occurrence frequencies in experimental segmental epitopes. These predictions were based on methods published by Kolaskar and Tongaonkar, Margalit and Spouge and Jameson and Wolf [26,27,28]. The experiments showed that the N′- and C′-terminus protein regions are mostly solvent accessible and unstructured, and an alpha-helical domain is the receptor-binding site; thus, we selected these regions of the PspA peptide sequence [29]. Antibody responses to such protein regions are likely to be recognized by antibodies raised against the native protein. The predicted antigenic peptides were analyzed in silico, and three prominent sequences were selected for the cloning and expression and antigenic property analysis via Western blotting.
Active Domain Fusion, Cloning, Expression and Western Blotting
In-silico determined/selected sequences were merged/fused together (189 base pairs; what we will call the fusion gene was synthesized using GenScript (GenScript, Inc., USA) and cloned in pTrcHisA expression vector (Invitrogen™, USA) for a pilot expression study in E. coli (Fig. 6). Fusion gene transformants were harvested and processed for intracellular expressing proteins. Expression was checked with time course 1 mM/mL IPTG induction from 0 to 16 h. Western blotting was carried out using a nitrocellulose membrane treated with a primary antibody [goat polyclonal Anti-PspA (bf-19; catalog no. sc17483)] using 1:500 dilution; secondary antibody, donkey anti-goat IgG-HRP (catalog no. sc2020), dilution 1:10,000. The nitrocellulose membranes were treated with the chemiluminescent western blot detection method (Thermo Scientific™ SuperSignal™ West Pico Chemiluminescent Substrate, catalog no. 34079) exposing the blot to the imaging system (radiography) for 30 s to 3 min, and the results were retrieved on X-ray film [30].
Results
Antibiotic Susceptibility of S. Pneumoniae Isolates
Susceptibility of isolated S. pneumoniae strains to 14 antibiotics commonly prescribing in Pakistani hospitals was evaluated. All isolated pneumococcal strains were susceptible to chloramphenicol and linezolid (MIC ≥ 0.5–1 μg/ml each); they showed responses to ampicillin (60%, MIC ≥ 0.06–1 μg/ml), amikacin (55%, MIC ≥ 1 μg/ml), amoxycillin (90%, MIC ≥ 0.25–1.5 μg/ml), co-amoxiclav (66%, MIC ≥ 0.06–0.12 μg/ml), cefotaxime (65%, MIC ≥ 0.06–0.25 μg/ml), ceftriaxone (60%, MIC ≥ 0.06–0.5 μg/ml), cefuroxime (60%, MIC ≥ 0.06–0.5 μg/ml), cephalexin (75%, MIC ≥ 1–2 μg/ml), fusidic acid (82%, MIC ≥ 0.12–1 μg/ml), teicoplanin (83%, MIC ≥ 0.03–0.12 μg/ml) and vancomycin (83%, MIC ≥ 0.12–1 μg/mL), while 20% of strains presented resistance to gentamycin (MIC ≥ 4–8 μg/ml) (Fig. 1).
Susceptibility testing of antibiotics. Strains (N = 38) tested with antibiotics commonly prescribed in Pakistani hospitals are shown on the X-axis following the Oxoid™, UK and EUCAST recommendations. The susceptibility pattern showed that all the strains were prone to chloramphenicol and linezolid, while 20% of strains presented resistance to gentamycin
Serotype Distribution of S. Pneumoniae Strains
The prevalence of pneumococcal serotypes obtained with the quellung reaction was as follows: strains belonging to serotype 19F, 23.60%; serotype 18B strains (non-vaccine type), 15.78%; 23F, 13.15%; 14, 10.52%; 10A, 10.52%; 3, 5.26%; 18C, 5.26%; 6B, 5.26%; 23B, (non-vaccine type), 2.63%; 6C (non-vaccine type), 2.63%; 8, 2.63%; serotype 12F, 2.63% (Fig. 2). No other serotypes or non-typeable strains were observed in the isolated pneumococcal strains (N = 38). The prevalence of serotypes included in PCV-7, PCV-10 or PCV-13 were 57.89, 57.89 and 73.68%, respectively, while serotypes included in PPSV-23 represented 89.47% of the experimental isolates (Fig. 2).
Percentage prevalence of pneumococcal serotypes isolated from pneumococcal patients (N = 38). Pneumococcal strains were serotyped by quellung reaction using the SSI® Pneumotest kit (Statens Serum Institut, Denmark). Serotype 19F was the leading type (23.60%), followed by 18B (15.78%). In the given number of samples, the exposure in PCV-7, PCV-10 and PCV-13 was 57.89, 57.89 and 73.68%, respectively, while serotypes included in PPSV23 comprised 89.47%
Amplification of the pspA Gene and Molecular Typing of Its Different Families and Clades
As the pspA gene is encoded by all pneumococcal strains, we amplified it using PCR from genomic DNA of all strains. To further classify the pspA gene in its different families, another set of PCR reactions was performed. These reactions utilized three different sets of family-specific primers and classified the pspA gene in family 1, 2 or 3. Our results demonstrated that most strains (N = 31) encoded a pspA gene belonging to family 2, whereas the remaining isolates (N = 7) were classified in family 1, and none were identified as family 3 (Fig. 3). These results revealed that PspA family 2 is the most prevalent form (~ 80%) of the PspA protein among Pakistani pneumococcal isolates and therefore it was our focus. The pspA genes belonging to family 2 were sequenced and distributed into their clades. All the sequences corresponding to the pspA gene family 2 were submitted to the NCBI GenBank and are available online under accession numbers KP337453, KU961865, KY010667 and KU961866 through KU961894.
Multiple Alignments and Phylogenetic Analysis
Nucleotide sequences of pspA genes and their amino acid sequences were examined with ClustalW2 and MEGA-6 software. Gene sequences obtained from Pakistani pneumococcal isolates were compared against the PspA native protein. Our analyses demonstrated that PspAs of N = 30 were the clade 3 genotype and N = 1 represented clade 4. The phylogenetic analyses showed that the PspA from a pneumococcal strain isolated in China (FJ663048.1) appears to be an ancestor of the Pakistani PspA family 2 protein (Fig. 4). Thus, whereas the PspA proteins produced by strains worldwide appear to be similar, our studies established high PspA sequence homologies among Pakistani isolates, different from those produced in other regions of the world.
The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 15.14 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6
Three-dimensional Structure and Antigenic Site Determination
Since we were interested in finding PspA-derivative antigenic peptides, our next study predicted the 3D structures of PspA, utilizing the DNAstar™ software. Based on the predicted motifs, we selected a 3D structure of PspA family 2 to further identify the antigenic epitopes that were most exposed. Three different methods, the (1) Kolaskar and Tongaonkar, (2) Margalit and Spouge and (3) Jameson and Wolf, were utilized to identify these epitopes. We selected three peptides in the PspA family 2 protein based on their characteristics, i.e., location, surface exposure, antigenicity, disordered ratio and hydrophilicity (Fig. 5). These peptides were analyzed in silico, and their properties are shown in Table 2. The selected peptides were primarily based on the N′- and C′-terminus regions and an alpha-helical domain of PspA. It was proposed that the PspA native protein is likely to be recognized by antibodies raised against any of these three selected peptides.
Peptides Fusion, Expression and Western Blotting
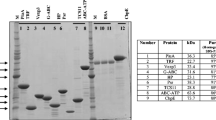
The selected three PspA family 2 segments were fused together into a fusion gene by the head-to-tail method and cloned in an expression vector pTrcHisA (Invitrogen™, USA) (Fig. 6). The DNA sequencing result showed that the fusion sequence is consistent with the designed one and with a correct open reading frame. The recombinant pTrcHisA vector containing the fusion gene was successfully transformed into competent E. coli for the pilot expression study and induced by 1 mM/ml IPTG. The SDS-PAGE and western blotting of bacterium lysate (processed with B-PER reagents) showed that the fusion peptide was successfully expressed. After 3 h induction, the fusion peptide reached the maximum in the time optimization test, although it was not expressed at 0 h induction. In western blotting, anti-PspA antibodies responded against this proposed fusion peptide like they did for the whole PspA protein (Fig. 7).
X-ray film radiography image (exposed for 3 min) of western blotting for PspA-fusion peptides ~ 10 kDa. Western blotting was carried out using nitrocellulose membranes treated with primary [goat polyclonal Anti-PspA (bf-19; catalog no. sc17483)] and secondary antibodies [donkey anti-goat IgG-HRP (catalog no. sc2020)]; 0–3 h was the 1 mM/ml IPTG induction time
Discussion
Antibiotics play a crucial role in the proper treatment of pneumococcal infections [31]. To the best of our knowledge, no studies showing the antibiotic-resistant profiles in the Lahore, Pakistan area are available. In the present study, we found that all isolated pneumococcal strains were susceptible to chloramphenicol and linezolid, while strains susceptible to gentamycin represented 20% of all cases (Fig. 1). The overall resistance of isolated pneumococcal strains to antibiotics in the Lahore, Pakistan hospital is low compared with other studies in other geographical regions. There is no direct association between susceptibility to antibiotics and serotypes. These data provide a baseline for the pneumococcal serotypes responsible for IPD cases and susceptibility or resistance to antibiotics in Pakistan.
Pakistan introduced PCV-7 in 2006 through the generous support of the Global Alliance for Vaccines and Immunizations (GAVI), and PCV-10 was being used in the country by 2013. However, the limited availability in hospitals and lack of awareness among the people in Pakistan, reduce its value. These are important issues for overall vaccine coverage in the country.
Our current study showed that most strains belonged to a serotype targeted by PCV13 (~ 73%), PCV7 (~ 57.89%) or PCV10 (~ 57.89%). This information is important given that most pneumococcal strains were isolated from PD cases from infants younger than 12 months of age and most of them would be protected if vaccinated with PCV13. However, PPSV23 has ~ 89.47% exposure in the Pakistani population according to our data. These data show that targeting PPSV23 to a large proportion of PD in the region is promising. An important and dangerous issue is that the 18B (non-vaccine type), 23B (non-vaccine type) and 6C (non-vaccine type) serotypes are also circulating in the Pakistani population (Fig. 2). The serotype distribution differs from a previous report shown by another group in the country [32]. This comparison might be related to temporal and regional differences and the potential circulation of outbreak strains. The work presented in this article is based on the most prevalent form of PD, pneumococcal pneumonia, with a few meningitis episodes. Data should therefore be interpreted keeping these important differences in mind.
The PspA-based vaccine for S. pneumoniae is an area requiring continuous investigation and development. Studies have shown that PspA has the potential for development into a human PspA-based peptide vaccine [33]. Reports available from the USA, Canada, France, Spain, Sweden, Australia, Japan and China have described that PspA family 2 is the most common among pneumococci [34,35,36,37,38]. The characterization of PspA presented in this study demonstrated that most pneumococcal strains isolated in Pakistan carried pspA genes, encoding the PspA protein belonging to family 2. A more extensive study might be required to draw complete conclusions. Our group will continue this study, especially quantitatively and in the other geographical areas of the country.
Various research groups have worked on different fragments of PspA and are claiming the antibodies are inducible [39, 40]. Western blot analysis revealed that there was strong recognition of the PspA fusion peptide. These results confirm that, among the PspA fragments analyzed in the literature [39, 40], our retrieved PspA family 2 segments would be the most suitable for the induction of broad reactivity, and this will be an even more effective strategy.
While it is essential to define which PspA family 2 fragment shows antigenicity within the same family, for this we will need to have longer fragments or include a macromolecular carrier. This could be a promising immunization strategy.
In general, our results showed that the PspA family 2 fusion peptide might be able to enhance the level of cross-reactivity in-vivo and will achieve cross-reactivity and have a broader effect. Moreover, future studies should also evaluate whether the protective ability of the antibodies correlates with the cross-reactivity data presented in this work. Our results will help future research regarding PspA-based peptide vaccines with broad specificity and coverage.
Previous studies have publicized that PspA families and clades are independent of serotypes. Pneumococci of different serotypes could be identified as the same PspA family/clade, and the same serotype could be identified in different families and/or clades [34,35,36,37,38]. Our data supported that every isolated pneumococcal strain encoded a pspA gene, its family and clade is independent of the serotype, and there is no correlation with antibiotic susceptibility (supplementary data provided). Thus, a PspA-based pneumococcal vaccine would have the potential to reduce the burden of PD in the area, possibly worldwide, with the advantage of avoiding the serotype replacement or shift observed with other pneumococcal vaccines.
Limitations of the Current Study
The data presented, pneumococcal serotypes 18B, 23B and 6C (non-vaccine types), which are not included in the current approved pneumococcal vaccine, but they are circulating in some ratio. It might be possible that some other non-vaccine types are also present in the region. Pneumococcal surface protein A family 2 is most prevalent in Pakistani pneumococcal isolates. For this, a more extensive study is required to reach definite conclusions. The PspA fusion peptide certainly revealed antigenic recognition in Western blotting, but it must be analyzed in an animal model for immunogenetic study.
Conclusion
The present introductory study reported that serotypes 19F and 18B (non-vaccine type) are the most prevalent serotypes isolated from PD cases in Pakistan. The prevalent PspA proteins produced by Pakistani isolates belong to family 2 and clade 3. They have high sequence homologies with each other and are different from those produced by strains isolated in the rest of the world. The PspA fusion peptide showed a positive antigenic response on Western blotting (Fig. 7), which will be further characterized in the recommended assays and an animal model (in vivo).
Change history
29 March 2018
In the original publication, one of the author names was missed in the author group
References
Tan TQ. Pediatric invasive pneumococcal disease in the United States in the era of pneumococcal conjugate vaccines. Clin Microbiol Rev. 2012;25(3):409–19.
O’Brien KL, Wolfson LJ, Watt JP, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374(9693):893–902.
Bogaert D, de Groot R, Hermans P. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4(3):144–54.
Calix JJ, Nahm MH. A new pneumococcal serotype, 11E, has a variably inactivated wcjE gene. J Infect Dis. 2010;202(1):29–38.
An Q, Hernandez A, Prejean J, German EJ, Thompson H, Hall HI. Geographic differences in HIV infection among Hispanics or Latinos—46 states and Puerto Rico, 2010. MMWR Morb Mortal Wkly Rep. 2012;61:805–10.
Defrance T, Taillardet M, Genestier L. T cell-independent B cell memory. Curr Opin Immunol. 2011;23(3):330–6.
Rennels MB, Edwards KM, Keyserling HL, et al. Safety and immunogenicity of heptavalent pneumococcal vaccine conjugated to CRM197 in United States infants. Pediatrics. 1998;101(4):604–11.
Darrieux M, Goulart C, Briles D, Leite LC. Current status and perspectives on protein-based pneumococcal vaccines. Crit Rev Microbiol. 2015;41(2):190–200.
Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011;378(9807):1962–73.
Mann B, Thornton J, Heath R, et al. Broadly protective protein-based pneumococcal vaccine composed of pneumolysin toxoid-CbpA peptide recombinant fusion protein. J Infect Dis. 2014;209(7):1116–25.
Hakansson A, Roche H, Mirza S, McDaniel LS, Brooks-Walter A, Briles DE. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect Immun. 2001;69(5):3372–81.
Tu AH, Fulgham RL, McCrory MA, Briles DE, Szalai AJ. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67(9):4720–4.
Crain MJ, Waltman WD 2nd, Turner JS, et al. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun. 1990;58(10):3293–9.
Ferreira DM, Darrieux M, Silva DA, et al. Characterization of protective mucosal and systemic immune responses elicited by pneumococcal surface protein PspA and PspC nasal vaccines against a respiratory pneumococcal challenge in mice. Clin Vaccine Immunol. 2009;16(5):636–45.
Hollingshead SK, Becker R, Briles DE. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun. 2000;68(10):5889–900.
Owais A, Tikmani SS, Sultana S, et al. Incidence of pneumonia, bacteremia, and invasive pneumococcal disease in Pakistani children. Trop Med Int Health. 2010;15(9):1029–36.
Nagaraj S, Kalal BS, Manoharan A, Shet A. Streptococcus pneumoniae serotype prevalence and antibiotic resistance among young children with invasive pneumococcal disease: experience from a tertiary care center in South India. Germs. 2017;7(2):78.
Castillo D, Harcourt B, Hatcher C, Jackson M, Katz L, Mair R. Identification and characterization of Streptococcus pneumoniae. Laboratory methods for the diagnosis of meningitis caused by neisseria meningitidis, Streptococcus pneumoniae, and haemophilus influenzae. 2011:73–86.
Chan WW, Lovgren M, Tyrrell GJ. Utility of the pneumoslide test in identification of Streptococcus pneumoniae in blood cultures and its relation to capsular serotype. J Clin Microbiol. 2009;47(5):1559–61.
Seki M, Yamashita Y, Torigoe H, Tsuda H, Sato S, Maeno M. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J Clin Microbiol. 2005;43(4):1581–6.
Slotved H-C, Kaltoft M, Skovsted I, Kerrn M, Espersen F. Simple, rapid latex agglutination test for serotyping of pneumococci (Pneumotest-Latex). J Clin Microbiol. 2004;42(6):2518–22.
Matuschek E, Brown DF, Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect. 2014;20(4):O255–66.
Li KB. ClustalW-MPI: clustalW analysis using distributed and parallel computing. Bioinformatics. 2003;19(12):1585–6.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–5.
Jameson B, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci CABIOS. 1988;4(1):181–6.
Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276(1–2):172–4.
Margalit H, Spouge JL, Cornette JL, Cease KB, Delisi C, Berzofsky JA. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J Immunol. 1987;138(7):2213–29.
Faraggi E, Xue B, Zhou Y. Improving the prediction accuracy of residue solvent accessibility and real-value backbone torsion angles of proteins by guided-learning through a two-layer neural network. Proteins. 2009;74(4):847–56.
Dawson NJ, Biggar KK, Storey KB. Characterization of fructose-1, 6-bisphosphate aldolase during anoxia in the tolerant turtle, Trachemys scripta elegans: an assessment of enzyme activity, expression and structure. PLoS ONE. 2013;8(7):e68830.
Song JH, Jung SI, Ko KS, et al. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob Agents Chemother. 2004;48(6):2101–7.
Mastro T, Spika J, Facklam R, et al. Antimicrobial resistance of pneumococci in children with acute lower respiratory tract infection in Pakistan. Lancet. 1991;337(8734):156–9.
Briles DE, Hollingshead S, Brooks-Walter A, et al. The potential to use PspA and other pneumococcal proteins to elicit protection against pneumococcal infection. Vaccine. 2000;18(16):1707–11.
Hollingshead SK, Baril L, Ferro S, et al. Pneumococcal surface protein A (PspA) family distribution among clinical isolates from adults over 50 years of age collected in seven countries. J Med Microbiol. 2006;55(Pt 2):215–21.
Hotomi M, Togawa A, Kono M, et al. PspA family distribution, antimicrobial resistance and serotype of Streptococcus pneumoniae isolated from upper respiratory tract infections in Japan. PLoS ONE. 2013;8(3):e58124.
Pimenta FC, Ribeiro-Dias F, Brandileone MC, Miyaji EN, Leite LC, Sgambatti de Andrade AL. Genetic diversity of PspA types among nasopharyngeal isolates collected during an ongoing surveillance study of children in Brazil. J Clin Microbiol. 2006;44(8):2838–43.
Qian J, Yao K, Xue L, et al. Diversity of pneumococcal surface protein A (PspA) and relation to sequence typing in Streptococcus pneumoniae causing invasive disease in Chinese children. Eur J Clin Microbiol Infect Dis. 2012;31(3):217–23.
Rolo D, Ardanuy C, Fleites A, Martin R, Linares J. Diversity of pneumococcal surface protein A (PspA) among prevalent clones in Spain. BMC Microbiol. 2009;9:80.
Moreno AT, Oliveira MLS, Ferreira DM, et al. Immunization of mice with single PspA fragments induces antibodies capable of mediating complement deposition on different pneumococcal strains and cross-protection. Clin Vaccine Immunol. 2010;17(3):439–46.
Darrieux M, Moreno AT, Ferreira DM, et al. Recognition of pneumococcal isolates by antisera raised against PspA fragments from different clades. J Med Microbiol. 2008;57(3):273–8.
Acknowledgements
We thank the Ethics Committee of the Children’s Hospital Lahore for providing permission to collect pneumococcal samples.
Funding
We acknowledge the support/funding from the Higher Education Commission (HEC) of Pakistan for the doctoral thesis (grant 2BM1-227). No funding or sponsorship was received for the publication of this article.
Authorship
All authors meet the criteria of the International Committee of Medical Journal Editors (ICMJE) for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval for the version to be published.
Disclosures
Faidad Khan, Mohsin Ahmad Khan, Nadeem Ahmed, Muhammad Islam Khan, Hamid Bashir, Saad Tahir and Ahmad Usman Zafar having nothing to disclose.
Compliance with Ethics Guidelines
The current study (both the study and collection of specimens) was approved by the Advanced Studies and Research Board of the University of the Punjab, Lahore, Pakistan (D/4439-ACAD), and sample collection from patients was approved by the Institutional Review Board (IRB)/Ethics Committee of the Children’s Hospital and Institute of Child Health, Lahore, Pakistan (CH.AD251). Specimens were collected (according to the recommended andapproved procedure) from these patients or children whose parents or guardians, agreed to participate in this study. It was specified that biologic material would only be utilized for research purposes. It is further clarified that all procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. All subjects provided written informed consent to participate in this study. An independent ethics committee or institutional review board reviewed the study protocol and amendments.
Thanking Patients
All the authors thank the patients and their families/guardians.
Data Availability
The data produced or evaluated during the current study and more information about the data are available from the corresponding author on reasonable request.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced content
To view enhanced content for this article go to https://doi.org/10.6084/m9.figshare.5909134.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Khan, F., Khan, M.A., Ahmed, N. et al. Molecular Characterization of Pneumococcal Surface Protein A (PspA), Serotype Distribution and Antibiotic Susceptibility of Streptococcus pneumoniae Strains Isolated from Pakistan. Infect Dis Ther 7, 277–289 (2018). https://doi.org/10.1007/s40121-018-0195-0
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-018-0195-0