Abstract
In this study, crude extracellular keratinase obtained from a novel keratin-degrading bacterial strain, Bacillus safensis LAU 13 (GenBank accession No. KJ461434) was used for the synthesis of silver nanoparticles (AgNPs). The particles were characterised by UV–Visible spectroscopy, Fourier transform infrared (FTIR) spectroscopy, and transmission electron microscopy. The biosynthesised AgNPs exhibited maximum absorbance at 409 nm. They are spherical in shape with the size ranging 5–30 nm. The FTIR spectrum showed peaks at 3410, 2930, 1664, 1618, 1389 and 600 cm−1, indicating that proteins were the capping and stabilisation molecules in the synthesis of AgNPs. Data obtained from XRD showed that the particles have face-centred cubic phase and are crystalline in nature with average size of ~8.3 nm. The particles showed effective inhibitory activity against five clinical isolates of Escherichia coli. Therefore, the keratinase of this strain could be used to develop an environmental friendly method for the rapid synthesis of AgNPs. To the best of our knowledge, this is the first report of green synthesis of AgNPs using the metabolite of B. safensis, and the report adds to the growing relevance of B. safensis as a potential industrially viable organism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nanobiotechnology is an offspring of nanotechnology that has emerged at the interface of nanotechnology and biology [1]. It is the multidisciplinary integration of biotechnology, nanotechnology, chemical processing, physical methodology and system engineering into biochips, molecular motors, nanocrystals and nanobiomaterials [2]. Nanotechnology deals with the synthesis and stabilisation of various nanoparticles [3]. There are different types of nanoparticles, but silver nanoparticles (AgNPs) have proved to be most effective with good antimicrobial efficacy [4]. AgNPs can be synthesised by using a variety of methods of synthesis like heating technique [5], laser irradiation [6], ionizing radiation [7], and radiolysis [8]. These methods are expensive and also hazardous to the environment. As a result, biological method of production is an alternative method, which has shown a very promising solution to these problems [9]. AgNPs have been successfully synthesised using plant extracts [10], bacteria [11], and fungi [12]. Biosynthesis of AgNPs is a bottom-up approach that mostly involves reduction/oxidation reactions [9]. In this way, microbial enzymes and phytochemicals with reducing potentials have been found responsible for the capping and stabilisation of nanoparticles. This is achieved in an eco-friendly manner, which does not lead to the use of toxic chemicals.
AgNPs have been used in different ways; such as in the treatment of burns, as dental materials, in textile fabrics, for water treatment, and as sunscreen lotions [13]. They also have applications in the production of antimicrobial paint [14], non-linear optics, spectrally selective coating for solar energy absorption and intercalation materials for electrical batteries, optical receptors, catalysis in chemical reactions, bio-labelling and antibacterial agents [15].
In our laboratory, we have recently isolated a novel strain of Bacillus safensis with potent keratinolytic activities (KA) [16] in a study for the biotechnological management of feather wastes and creation of novel products. The aim of this study was to investigate the ability of crude extracellular keratinase of this bacterial strain for the production of AgNPs and evaluation of its applications. It is an attempt to extend the frontier of the organism for biotechnological applications.
Materials and methods
Microorganism
The bacterium used in this study was isolated following the methods described by Lateef et al. [17]. The medium used for isolation consisted of the following in g/L: NaNO3, 2; NaCl, 2; KH2PO4, 2; MgSO4, 0.05; FeSO4.7H2O, 0.1; CaCO3, 0.1; feather powder, 20; and agar–agar, 20 (BDH, UK). The pure isolate was identified using biochemical methods [18], and 16S rRNA analysis. Genomic DNA of the bacterium was isolated as described by Sambrook et al. [19], and the DNA fragments were amplified using polymerase chain reaction (PCR) technique at 68–72 °C with BacF (5′-GGGAAACCGGGGCTAATACCGGAT-3′) and R1378 (5′-CGGTGTGTACAAGGCCCGGGAACG-3′) primers which are the reverse and forward primers, respectively. From single colonies of the isolate, two replicates of the 16S rRNA gene (approximately 1,500 base pairs) were sequenced using Sanger’s method. These sequences were used in the database of National Centre for Biotechnological Information (NCBI) (BLAST, http://blast.ncbi.nlm.nih.gov/) via BLASTN option [20] for possible matches, and thereafter submitted to GenBank for accession number.
Keratinase production
Inoculum was developed by inoculating a loopful of pure culture into a medium consisting of 1 % keratin substrate and 0.2 % yeast extract (pH 7.50). The culture was incubated at 37 °C and 100 rpm for 24 h. About of 1 ml of inoculum was inoculated into 19 ml/100 ml flask of liquid minimal medium, and the flasks were incubated at 37 °C at 100 rpm for 120 h. At 24 h intervals, whole flasks were taken out, the broth centrifuged at 5,000 rpm at 10 °C for 20 min, and the supernatant served as crude extracellular keratinase.
Keratinase assay
The keratinase activity was determined by the modified method of Cheng et al. [21]. About 0.5 ml of crude enzyme was incubated with 1.5 g of feather powder suspended in 2 ml of phosphate buffer (pH 7.5). The control experiment consisted of buffer and feather powder only. The reaction mixtures were incubated at 40 °C for 3 h at 100 rpm. The reaction was terminated by adding 2 ml of 10 % trichloroacetic acid (TCA). The reaction mixture was centrifuged at 5,000 rpm for 15 min at room temperature (30 ± 2 °C) to remove precipitated keratin. The increase in absorbance at 280 nm of the filtrate of the test sample relative to that of the control, a measure of release of protein was converted into keratinase units (1U = 0.01 absorbance increase for 1 h reaction time [22]), was measured.
Synthesis and characterisation of silver nanoparticles (AgNPs)
AgNPs were synthesised by reacting the crude keratinase with silver nitrate solution as described by Revathi et al. [23]. About 1 ml of crude enzyme (50 U ml−1) was added to the reaction vessel containing 50 ml of 1 mM silver nitrate (AgNO3) solution for the reduction of silver ion. The reaction was carried out in static condition at room temperature (30 ± 2 °C) for 2 h. The formation of AgNPs was monitored through visual observation of the change of colour, and measurement of the absorbance spectrum of the reaction mixture using UV–Visible spectrophotometer (Genesys 10 UV Thermoelectron Corporation, UK).
Fourier transform infrared (FTIR) spectroscopy analysis was carried out using BUCK M530 Spectrophotometer (Buck, USA) on the powder sample of AgNPs according to Bhat et al. [1]. The AgNPs solution was centrifuged at 10,000 rpm for 20 min. The solid residue obtained was then dried at room temperature, and the powder obtained was used for FTIR measurement using KBr pellets. The X-ray diffraction (XRD) analysis was investigated using PANalytical X’pert PRO with CuK α radiation at 40 kV and 30 mA.
The transmission electron microscopy (TEM) micrograph was obtained as follows: A drop of nanoparticles in suspension was placed on a 200 mesh hexagonal copper grid (3.05 mm) (Agar Scientific, Essex, UK) coated with 0.3 % formvar dissolved in chloroform. The particles were allowed to settle for 3–5 min on the grid, the excess liquid flicked off with a wick of filter paper and the grids were then air dried before TEM viewing. Micrograph was obtained using a JEM-1400 (JEOL, USA) operating at 200 kV.
Antibacterial activity of synthesised AgNPs
The antibacterial activity of the AgNPs was examined using disc diffusion method as described by Thillaimaharami et al. [24]. This was carried out using 6 mm diameter paper disc prepared from Whatman No. 1 filter paper. A loopful of each of five Escherichia coli strains isolated from wound was suspended in 10 ml of sterile distilled water. This was used to make subsequent tenfold dilutions from which 0.1 ml was transferred into 4 ml of sterilised nutrient agar already cooled to approximately 50 °C, which was poured over a nutrient agar base plate and allowed to solidify. The paper discs were impregnated with AgNPs solutions that were obtained by serial dilution with distilled water (150 µg ml−1). These were placed on the inoculated nutrient agar plates, while the control disc was impregnated with only crude keratinase. The zone of inhibition was determined after incubation at 37 °C for 24 h.
Results and discussion
Bacterial isolate
Four bacterial strains were isolated from the screened feather dumping site amongst which B. safensis LAU 13 (GenBank accession No KJ461434) produced highest keratinolytic activity [16]. It is a rod-shaped, Gram-positive, spore-forming bacterium. The analysis based on 16S rRNA sequence showed that this strain has 100 % sequence homology with B. safensis CBN-8 (JQ353775). Bacillussafensis was first identified in 2006 as a contaminant from spacecraft-assembly facilities (SAF) in USA from which it derived its specific epithet safensis [25]. Bacillus safensis are salt-tolerating plant growth promoting rhizobacteria (PGPR) [26, 27]. The productions of industrially important enzymes like β-galactosidase [28], endoinulinase [29], and lipases [30] by some strains of B. safensis have been reported. In addition, we recently established the first report of the production of keratinase by a strain of B. safensis [16]. However, there is no report on the utilisation of the bacterium for the synthesis of nanoparticles. Therefore, the production of nanoparticles using keratinase of B. safensis adds to the potential relevance of the bacterium for industrial applications.
Keratinolytic activities during the growth of B. safensis LAU 13
The strain produced KA in the range 35.4–50.4 U ml−1 during 120 h of cultivation using feather as keratin substrate. The enzyme activity reached a maximum of 50.4 U ml−1 at 72 h of cultivation, after which it decreased to a final value of 36.7 U ml−1 at 120 h, which is similar to KA reported for some bacilli [17, 31].
Biosynthesis and characterisation of silver nanoparticles
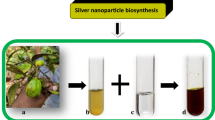
There is limited report on the use of keratinases for the production of nanoparticles, as only a single report of such exists [23]. In this study, crude extracellular keratinase from B. safensis LAU 13 was used to synthesise AgNPs. The AgNPs were characterised with colour formation produced from the reduction of silver ion by the enzyme. The intensity of colour increases as the bio-reduction of silver ion progresses and becomes stable when the reaction is completed. Figure 1 shows the dark brown solution formed from the reaction of crude keratinase and AgNO3 solution after 1.5 h of reaction, while the AgNO3 solution in the control experiment remained unchanged. The dark brown solution formed is an indication of the synthesis of AgNPs.
The colour of biosynthesised AgNPs reported by some authors varies, which may be attributed to the variations in the composition of biomolecules. Kalishwaralal et al. [32] reported AgNPs of brown colour synthesised from culture supernatant of Bacillus licheniformis, while Shaligram et al. [3] reported dark brown-coloured AgNPs synthesised from microbial culture supernatant. In a recent study, Revathi et al. [23] reported the synthesis of dark brown-coloured AgNPs from crude keratinases after 72 h of incubation. These colour formations are due to the excitation of surface plasmon vibrations in metal nanoparticles [33]. It was observed in this study that the crude keratinase from this strain can rapidly react with silver nitrate solution to form AgNPs.
Figure 2a shows the UV–Visible spectra of the biosynthesised AgNPs in the range 190–1,100 nm and the absorption spectrum peaked at 409 nm. The AgNPs gave a clear solution when dispersed in distilled water for extended period of up to 6 months, with the characteristic absorption spectrum peak (Fig. 2b), which is an indication of the small sizes of the particles. This characteristic absorption peak justified the formation of AgNPs. Several authors have reported peaks of absorption spectra of AgNPs in the range 391–440 nm [10, 11, 34–37], while the peak at 292 nm could be attributed to the presence of proteins [38]. However, plasmon resonances for the transverse and longitudinal plasmon bands at 350 and 390 nm, respectively, typified of anisotropic metallic one-dimensional nanostructures have been reported [39].
The FTIR measurements were carried out to identify the possible biomolecules responsible for the stabilisation of the synthesised AgNPs. The FTIR spectrum (Fig. 3) showed peaks at 3410, 2930, 1664, 1618, 1389 and 600 cm−1. The peak of 2,930 cm−1 indicated secondary amine [40], while another peak at 1,664 cm−1 was recognised as amide I and had been previously shown to be responsible for the capping of silver ion [35]. It has been reported that proteins can bind to nanoparticles either through free amine groups or cysteine residues in the proteins [41], hence, it can be inferred that these biomolecules are responsible for capping and efficient stabilisation of the AgNPs.
The TEM micrograph of the AgNPs is as shown in Fig. 4. The AgNPs are spherical in shape and the size ranged 5–30 nm. Spherical shape of AgNPs has been reported [36], while Kannan et al. [10] reported the synthesis of AgNPs in the size range 3–44 nm. These properties may influence the activity of AgNPs especially antimicrobial activity. This study has shown the capability of keratinase of B. safensis at forming nanoparticles, which may add to the biofunctional and biotechnological usefulness of keratinases and B. safensis.
The XRD pattern of the synthesised AgNPs shows the presence of peaks at 2θ = 38°, 44°, 64° and 77° which correspond to (111), (200), (220) and (311) planes of silver, respectively (Fig. 5). The AgNPs are polycrystalline as revealed by the significant peak intensities of the crystal planes which are consistent with published studies [42, 43]. From the spectrum, existence of single peaks at different diffraction angles implies that the particles are of a homogeneous material without impurities. All the peaks in XRD pattern are readily indexed to a face-centred cubic structure of silver (JCPDS, File No. 4-0783). The crystallite size (L) of the nanoparticles is calculated from Scherrer’s formula:
where λ is the wavelength (0.15,418 Å) of X-rays used, β is the full width at half maximum (FWHM) measured in radians and θ is the Bragg’s angle. Using (111) plane, L is ~8.3 nm. Based on Bragg’s formula, the lattice spacing is calculated to be 0.236 nm while the lattice constant is estimated to be 0.4096 nm, which is consistent with the standard value of 0. 4086 nm.
; d(hkl) is the lattice spacing while a is the lattice parameter. The average particle size of ~8.3 nm as revealed by XRD further confirms the particle size data of 5–30 nm obtained through TEM.
Antibacterial activities of silver nanoparticles
The AgNPs showed remarkable antimicrobial activity against five clinical isolates of E.coli. The AgNPs induced a maximum zone of 12.5 mm on the test strains, while the minimum zone was 8.6 mm at concentration of 150 µg ml−1 (Table 1). The efficacies of biosynthesised AgNPs have been reported against E. coli [11]. Furthermore, AgNPs have been reported to have high surface area to volume ratio which leads to excellent antimicrobial activity as compared with bulk silver metal [44], because there will be close attachment of the nanoparticles surface with microbial cells, making its antimicrobial property to be size dependent [1]. Through the interaction of AgNPs with sulphur and phosphorus containing biomolecules in the bacterial cell, the particles enter into the cell, where cell-killing is initiated through the attack of the respiratory chain and cell division [44]. Therefore, the appreciable antibacterial property demonstrated by the AgNPs proved that it could either be mixed with antibiotic drugs or used directly as drugs for some topical applications.
Conclusion
This study has led to the green synthesis of AgNPs by crude extracellular keratinase obtained through the biotechnological management of feather waste using a novel strain of B.safensis. AgNPs of uniform spherical shape of ~8.3 nm and face-centred cubic structure have been produced. The FTIR analysis revealed that protein molecules were responsible for the stabilisation and capping of the AgNPs, and the particles also demonstrated excellent antimicrobial property. Therefore, the stabilised and uniform shaped AgNPs obtained from this easy and eco-friendly method of synthesis could be used in drug formulation for topical applications and also for various biotechnological applications. To the best of our knowledge, this is the first report of the use of the metabolite of B. safensis in the green synthesis of nanoparticles.
References
Bhat, R., Deshpande, R., Ganachari, S.V., Huh, D.O., Venkataraman, A.: Photo-irradiated bio-synthesis of silver nanoparticles using edible mushroom Pleurotus florida and their antibacterial activity studies. Bioinorg. Chem. Appl. 2011, 1–7 (2011)
Heinrich, K.: Nanotechnology: from molecules to systems. Eng. Life Sci. 4, 211–218 (2004)
Shaligram, N.S., Bule, M., Bhambure, R., Singhal, R.S., Singh, S.K., Szakacs, G., Pandey, A.: Biosynthesis of silver nanoparticles using aqueous extract from the compactin producing fungal strain. Process Biochem. 44, 939–943 (2009)
Gong, P., Li, H., He, X., Wang, K., Hu, J., Tan, W., et al.: Preparation and antibacterial activity of Fe3O4 and Ag nanoparticles. Nanotechnology 18, 604–611 (2007)
Huang, H., Yang, X.: Synthesis of polysaccharide-stabilized gold and silver nanoparticles: a green method. Carbohydr. Res. 339(15), 2627–2631 (2004)
Abid, J.P., Wark, A.W., Brevet, P.F., Girault, H.H.: Preparation of silver nanoparticles in solution from a silver salt by laser irradiation. Chem. Commun. 7, 792–793 (2002)
Gasaymeh, S.S., Radiman, S., Heng, L.Y., Saion, E., Saeed, H.M.: Synthesis and characterization of silver/polyvinilpirrolidone (AG/PVP) nanoparticles using Gamma irradiation techniques. Am. J. Appl. Sci. 7(7), 892–901 (2010)
Soroushian, B., Lampre, I., Belloni, J., Mostafavi, M.: Radiolysis of silver ion solutions in ethylene glycol: solvated electron and radical scavenging yields. Radiat. Phys. Chem. 72, 111–118 (2005)
Prabhu, S., Poulose, E.K.: Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int. Nano Lett. 2, 32 (2012)
Kannan, R.R.R., Arumugam, R., Ramya, D., Manivannan, K., Anantharaman, P.: Green synthesis of silver nanoparticles using marine macroalga Chaetomorpha linum. Appl. Nanosci. 3, 229–233 (2013)
Priyadarshini, S., Gopinath, V., Priyadharsshini, N.M., Ali, D.M., Velusamy, P.: Synthesis of anisotropic silver nanoparticles using novel strain, Bacillus flexus and its application. Colloids Surf. B 102, 232–237 (2013)
Selvi, K.V., Sivakumar, T.: Isolation and characterization of silver nanoparticles from Fusariumoxysporium. Int. J. Curr. Microbiol. Appl. Sci. 1(1), 56–62 (2012)
Duran, N., Marcarto, P.D., De Souza, G.I.H., Alves, O.L., Esposito, E.: Antibacterial effect of silver nanoparticles produced by fungal process on textile fabrics and their effluent treatment. J. Biomed. Nanotechnol. 3, 203–208 (2007)
Kumar, A., Vemula, P.K., Ajayan, P.M., John, G.: Silver-nanoparticle-embedded antimicrobial paints based on vegetable oil. Nat. Mater. 7(3), 236–241 (2008)
Nelson, D., Priscyla, D.M., Oswaldo, L.A., Gabriel, I.H., De Souza, E.E.: Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J. Nanobiotechnol. 3, 1–7 (2005)
Lateef, A., Adelere, I.A., Gueguim-Kana, E.B.: Bacillussafensis LAU 13: a new novel source of keratinase and its multi-functional biocatalytic applications. Biotechnol. Biotechnol. Equip. doi:10.1080/13102818.2014.986360 (in press)
Lateef, A., Oloke, J.K., Gueguim-Kana, E.B., Sobowale, B.O., Ajao, S.O., Bello, B.Y.: Keratinolytic activities of a new feather-degrading isolate of Bacillus cereus LAU 08 isolated from Nigerian soil. Int. Biodeterior. Biodegrad. 64, 162–165 (2010)
Brenner, D.J., Krieg, N.R., Staley, J.T.: Bergey’s Manual of Systematic Bacteriology. Part B, 2nd edn, pp. 323–358. Springer, New York (2004)
Sambrook, J., Fritch, E.F., Maniatis, T.: Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor (1989)
Cheng, S.W., Hu, H.M., Shen, S.W., Takagi, H., Asano, M., Tsai, Y.C.: Production and characterization of a feather degrading Bacillus licheniformis PWD-1. Biosci. Biotechnol. Biochem. 59, 2239–2243 (1995)
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J.: Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990)
Ramnani, P., Gupta, R.: Optimization of medium composition for keratinase production on feather by Bacillus licheniformis RG1 using statistical methods involving response surface methodology. Biotechnol. Appl. Biochem. 40, 491–496 (2004)
Revathi, K., Shaifali, S., Mohd, A.K., Suneetha, V.: A potential strain of keratinolytic bacteria VIT RSAS2 from katpadi and its pharmacological benefits. Int. J. Pharm. Sci. Res. 20(2), 89–92 (2013)
Thillaimaharani, K., Sharmila, K., Thangaraju, P., Karthick, M., Kalaiselvam, M.: Studies on antimicrobial and antioxidant properties of oyster mushroom Pleurotus florida. Int. J. Pharm. Sci. Res. 4, 1540–1545 (2013)
Satomi, M., Myron, T., Duc, L., Venkateswaran, K.: Bacillus safensis sp. nov., isolated from spacecraft and assembly-facility surfaces. Int. J. Syst. Evol. Microbiol. 56, 1735–1740 (2006)
Chakraborty, U., Chakraborty, B.N., Chakraborty, A.P.: Dey, PL Water stress amelioration and plant growth promotion in wheat plants by osmotic stress tolerant bacteria. World J. Microbiol. Biotechnol. 29(5), 789–803 (2012)
Kothari, V.V., Kothari, R.K., Kothari, C.R., Bhatt, V.D., Nathani, N.M., Koringa, P.G., Joshi, C.G., Vyas, B.R.M.: Genomic sequence of salt-tolerant Bacillus safensis strain VK, isolated from saline desert area of Gujarat, India. Genome A 1(5), 00671–00713 (2013)
Nath, A., Chakrabarty, S., Sarkar, S., Bhattacharjee, C., Drioli, E., Chowdhury, R.: Purification and characterization of β-galactosidase synthesized from Bacillussafensis (JUCHE 1). Ind. Eng. Chem. Res. 52(33), 11663–11672 (2013)
Singh, R.S., Singh, R.P., Yadav, M.: Molecular and biochemical characterization of a new endoinulinase producing bacterial strain of Bacillus safensis AS-08. Biologia 68(6), 1028–1033 (2013)
Kumar, D., Parshad, R., Gupta, V.K.: Application of a statistically enhanced, novel, organic solvent stable lipase from Bacillus safensis DVL-43. Int. J. Biol. Macromol. 66, 97–107 (2014)
Joshi, S.G., Tejashwini, M.M., Revati, N., Sridevi, R., Roma, D.: Isolation, identification and characterization of a feather degrading bacterium. Int. J. Poult. Sci. 6, 689–693 (2007)
Kalishwaralal, K., Deepak, V., Ramkumarpandian, S., Nellaiah, H., Sangiliyandi, G.: Extracellular biosynthesis of silver nanoparticles by the culture supernatant of Bacillus licheniformis. Mater. Lett. 62, 4411–4413 (2008)
Mulvaney, P.: Surface Plasmon spectroscopy of nanosized metal particles. Langmuir 12(3), 788–800 (1996)
Kalimuthu, K., Babu, R.S., Venkataraman, D., Bilal, M., Gurunathan, S.: Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf. B 65, 150–153 (2008)
Babu, M.M.G., Gunasekaran, P.: Production and structural characterization of crystalline silver nanoparticles from Bacillus cereus isolate. Colloids Surf. B 74, 191–195 (2009)
Zaki, S., El-Kady, M.F., Abd-El-Haleem, D.: Biosynthesis and structural characterization of silver nanoparticles from bacterial isolates. Mater. Res. Bull. 46, 1571–1576 (2011)
Thirumurugan, A., Tomy, N.A., Kumar, H.P., Prakash, P.: Biological synthesis of silver nanoparticles by Lantana camara leaf extracts. Int. J. Nanomater. Biostruct. 1(2), 22–24 (2011)
Bhainsa, K.C., D’Souza, S.F.: Extracellular biosynthesis of silver nanoparticles using the fungus Aspergillus fumigatus. Colloids Surf. B 47, 160–164 (2006)
Amirjani, A., Marashi, P., Fatmeshari, D.H.: The effects of physicochemical parameters on the synthesis of silver nanowires via polyol method. Int. Nano Lett. 4, 108 (2014)
Vigneshwaran, N., Kathe, A.A., Varadarajan, P.V., Nachane, R.P., Balasubramanya, R.H.: Biomimetics of silver nanoparticles by white rot fungus, Phaenerochaete chrysosporium. Colloids Surf. B Biointerfaces 53, 55–59 (2007)
Mandal, S., Phadtare, S., Sastry, M.: Interfacing biology with nanoparticles. Curr. Appl. Phys. 5, 118–127 (2005)
Sathishkumar, M., Sneha, K., Yun, Y.-S.: Immobilization of silver nanoparticles synthesized using Curcumalonga tuber powder and extract on cotton cloth for bactericidal activity. Bioresour. Technol. 101, 7958–7965 (2010)
Kanmani, P., Lim, S.T.: Synthesis and structural characterization of silver nanoparticles using bacterial exopolysaccharide and its antimicrobial activity against food and multidrug resistant pathogens. Process Biochem. 48, 1099–1106 (2013)
Mahendra, R., Alka, Y., Aniket, G.: Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 27(1), 76–83 (2009)
Acknowledgments
AL thanked authority of LAUTECH, Ogbomoso for providing fund for some aspects of this work through the TETFund Research Project Intervention (APU/TETFund/114) for the project titled ‘Efficient Biotechnological Management of Keratin Waste to create Novel Products’. AIA gratefully acknowledged authority of FUT, Minna for a postgraduate study leave.
Conflict of interest
There are no competing interests.
Authors’ contributions
AL designed the framework, led the investigation, interpreted the data and wrote the paper with AIA. AIA carried out the laboratory investigations and library resources, while GKEB, TBA and BLS carried out advanced analysis of the samples. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under license to BioMed Central Ltd. Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Lateef, A., Adelere, I.A., Gueguim-Kana, E.B. et al. Green synthesis of silver nanoparticles using keratinase obtained from a strain of Bacillus safensis LAU 13. Int Nano Lett 5, 29–35 (2015). https://doi.org/10.1007/s40089-014-0133-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40089-014-0133-4