Abstract
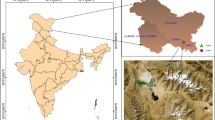
Heavy metal pollution poses a serious threat to soil, water, and the atmospheric environment. Many microbes sustain and respond to the metal stress, and the mechanisms involved in these processes remain unclear. The metagenomics and metatranscriptomics approaches were applied to study the structure and function of eukaryotic microbial communities in heavy metal-contaminated soils of two geographic locations situated in different climatic regions. To achieve this, amplicons of the hypervariable V4 region of 18S rDNA and cDNA synthesized from 18S rRNA extracted from these soils were generated and sequenced through paired-end sequencing chemistry on the Illumina-MiSeq platform. The NGS dataset processed by the Mothur pipeline and analyzed by Parallel-Meta 3 pipeline illustrated the presence of all the major eukaryotic phyla. Taxa diversity and community structure of micro-eukaryotes within and between the samples from two locations were compared. Clustering, heatmap, and PCA analysis supported the variation in taxonomic diversity and community structure in the datasets of these two sites. Analysis of taxa abundance in both sites identified marker organisms for the further characterization of such types of environments. A functional metatranscriptomics study revealed the identification of various expressed eukaryotic genes, which are involved in metal tolerance. These metal-tolerant gene families were phylogenetically related to the different eukaryotic lineages reported in the metagenomic analysis. Hence, this approach could be widely applied in microbial ecology to understand the role of active microbes in such specific environmental conditions.



Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within this article and the sequence information in the NCBI database.
References
Arkadeep Mukherjee M, Reddy S (2020) Metatranscriptomics: an approach for retrieving novel eukaryotic genes from polluted and related environments. 3 Biotech. https://doi.org/10.1007/s13205-020-2057-1
Bailly J, Fraissinet-Tachet L, Verner MC, Debaud JC, Lemaire M, Wésolowski-Louvel M et al (2007) Soil eukaryotic functional diversity, a metatranscriptomic approach. ISME J 1:632–642
Bates ST, Clemente JC, Flores GE, Walters WA, Parfrey LW, Knight R et al (2013) Global biogeography of highly diverse protistan communities in soil. ISME J 7:652–659
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y et al (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Damon C, Lehembre F, Oger-Desfeux C, Luis P, Ranger J, Fraissinet-Tachet L, Marmeisse R (2012) Metatranscriptomics reveals the diversity of genes expressed by eukaryotes in forest soils. PLoS ONE 7(1):e28967. https://doi.org/10.1371/journal.pone.0028967
De Vargas C, Audic S, Henry N, Decelle J, Mahe F, Lagos, (2015) Eukaryotic plankton diversity in the sunlight ocean. Science 348:1261605
Dixit AR, Dhankher OP (2011) A novel stress-associated protein ‘AtSAP10’from Arabidopsis thaliana confers tolerance to nickel, manganese, zinc, and high temperature stress. PLoS ONE 6:e20921
Eilers KG, Lauber CL, Knight R, Fierer N (2010) Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem 42:896–903
Eiler A, Drakare S, Bertilsson S, Pernthaler J, Peura S et al (2013) Unveiling distribution patterns of freshwater phytoplankton by a Next Generation Sequencing based approach. PLOS ONE 8:e53516
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70:603–647
Gans J, Wolinsky M, Dunbar J (2005) Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:31–34
Griffiths BS, Philippot L (2013) Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol Rev 37:112–129
Guo L, Sui Z, Zhang S, Ren Y, Liu Y (2015) Comparison of potential diatom “barcode” genes (18S and ITS rDNA, COI, rbcL) and their effectiveness in discriminating and determining species taxonomy in Bacillariophyta. Int J Syst Evol Microbiol 65:1369–1380
Hadziavdic K, Lekang K, Lanzen A (2014) Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 9:e87624
Hebert PD, Ratnasingham S, De Waard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc B 270:S96–S99
Hugerth LW, Muller EE, Hu YO, Lebrun LA, Roume H, Lundin D (2014) Systematic design of 18S rRNA gene primers for determining eukaryotic diversity in microbial consortia. PLoS ONE 9:e95567
Jing G, Sun Z, Wang H, Gong Y, Huang S, Ning K, Xu J (2017) Su X (2017) Parallel-META 3: Comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci Rep 12(7):40371
Jubeaux G, Audourd-Combe F, Simon R, Tutundijn R, Salvador A, Geffard O et al (2012) Vitellogenin- like proteins among invertebrate species diversity: potential of proteomic mass spectrometry for biomarker development. Environ Sci Technol 46:6315–6323
Kitson RE, Mellon MG (1954) Colorimetric determination of phosphorus as molybdivanadophosphoric acid. Ind Eng Chem 16:379–383
Lehembre F, Doillon D, David E, Perrotto S, Baude J, Foulon J et al (2013) Soil metatranscriptomics for mining eukaryotic heavy metal resistance genes. Environ Microbiol 15:2829–2840
Leimena MM, Ramiro-Garcia J, Davids M, van den Bogert B, Smidt H, Smid EJ et al (2013) A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genomics 14:530
Lesaulnier C, Papamichail D, McCorkle S, Ollivier B, Skiena S et al (2008) Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ Microbiol 10:926–941
Lohan KP, Fleischer RC, Carney KJ, Holzer KK, Ruiz GM (2016) Amplicon-based pyrosequencing reveals high diversity of protistan parasites in ships’ ballast water: implications for biogeography and infectious diseases. Microbial Ecol 71:530–542
Manoharan L, Kushwaha SK, Ahrén D, Hedlund K (2017) Agricultural land use determines functional genetic diversity of soil microbial communities. Soil Biol Biochem 115:423–432
Marmeisse R, Kellner H, Fraissinet-Tachet L, Luis P (2017) Discovering protein-coding genes from the environment: Time for the eukaryotes? Trends Biotechnol 35:824–835
Micó C, Recatalá L, Peris M, Sánchez J (2006) Assessing heavy metal sources in agricultural soils of a European Mediterranean area by multivariate analysis. Chemosphere 65:863–872
Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Curr Genet 2:417–422
Mukherjee A, Yadav R, Marmeisse R, Fraissinet-Tachet L, Reddy MS (2019a) Heavy metal hypertolerant eukaryotic aldehyde dehydrogenase isolated from metal contaminated soil by metatranscriptomics approach. Biochimie 160:183–192
Mukherjee A, Yadav R, Marmeisse R, Fraissinet-Tachet L, Reddy MS (2019b) Detoxification of toxic heavy metals by serine protease inhibitor isolated from polluted soil. Int Biodeterior Biodegr 143:104718
Mukherjee A, Thakur B, Pandey AK, Marmeisse R, Fraissinet-Tachet L, Reddy MS (2021) Multi-metal tolerance of DHHC palmitoyl transferase-like protein isolated from metal contaminated soil. Ecotoxicology 30:67–79
O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71:5544–5550
Olsen SR (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. United States Department of Agriculture, Washington
Piper CS (1966) Soil and plant analysis; A laboratory manual of methods for the examination of soils and the determination of the inorganic constitutents of plants. Hans Publishers; Bombay
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59
Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Brietbart M et al (2010) Viral and microbial community dynamics in four aquatic environments. ISME J 4:739
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR et al (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shitan N, Horiuchi KI, Sato F, Yazaki K (2007) Bowman-Birk proteinase inhibitor confers heavy metal and multiple drug tolerance in yeast. Plant Cell Physiol 48:193–197
Sobecky PA, Coombs JM (2009) Horizontal gene transfer in metal and radionuclide contaminated soils. Methods Mol Biol 532:455–472
Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner HW et al (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19:S21–S31
Suzuki N, Koizumi N, Sano H (2001) Screening of cadmium responsive genes in Arabidopsis thaliana. Plant Cell Environ 24:1177–1188
Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI et al (2014) Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci USA 111:6341–6346
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R (2014) Global diversity and geography of soil fungi. Science 346:1256688
Thakur B, Yadav R, Fraissinet-Tachet L, Mermeisse R, Reddy MS (2018) Isolation of multi-metal tolerant ubiquitin fusion protein from metal polluted soil by metatranscriptomic approach. J Microbiol Methods 152:119–125
Thakur B, Yadav R, Vallon L, Marmeisse R, Fraissinet-Tachet L, Reddy MS (2019) Multi-metal tolerance of von Willebrand factor type D domain isolated from metal contaminated site by metatranscriptomics approach. Sci Total Environ 661:432–440
Thakur B, Yadav R, Mukherjee A, Melayah D, Marmeisse R, Fraissinet-Tachet L et al (2021) Protection from metal toxicity by Hsp40-like protein isolated from contaminated soil using functional metagenomic approach. Environ Sci Pol Res 28:17132–17145
Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE 3:e2527
Vaulot D, Eikrem W, Viprey M, Moreau H (2008) The diversity of small eukaryotic phytoplankton in marine ecosystems. FEMS Microbiol Rev 32:795–820
Walkley A (1947) A critical examination of a rapid method for determining organic carbon in soils-effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci 63:251–264
Woo S, Yum S, Park HS, Lee TK, Ryu JC (2009) Effects of heavy metals on antioxidants and stress-responsive gene expression in Javanese medaka (Oryzias javanicus). Comp Biochem Physio. Part C: Toxicol Pharmacol 149:289–299
Yadav RK, Barbi F, Ziller A, Luis P, Marmeisse R, Reddy MS, Fraissinet-Tachet L (2014) Construction of sized eukaryotic cDNA libraries using low input of total environmental metatranscriptomic RNA. BMC Biotechnol 14:80–86
Zhang P, Qin C, Hong X, Kang G, Qin M, Yang D et al (2018) Risk assessment and source analysis of soil heavy metal pollution from lower reaches of Yellow River irrigation in China. Sci Total Environ 633:1136–1147
Acknowledgements
The authors are thankful to Indo-French Centre for the Promotion of Advanced Research, New Delhi, for financial support under project No. 4709-1.
Funding
This research received a grant from Indo-French Centre for the Promotion of Advanced Research, New Delhi, under project No. 4709–1.
Author information
Authors and Affiliations
Contributions
BT and RKY performed all the sequencing experiments. BT analyzed the NGS data, wrote the manuscript, and. PS and KMM helped in the acquisition of the data and contributed to the data analysis. MSR, RM, and LFT conceived the study and designed experiments and manuscript revision. MSR and LFT, the principal investigator for the grant and for constructing the experimental design, supervised the project and helped to write the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict Of interest
The authors declare no potential conflict of interest.
Additional information
Editorial responsibility: Samareh Mirkia.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thakur, B., Yadav, R.K., Marmeisse, R. et al. Metagenomic analysis of heavy metal-contaminated soils reveals distinct clades with adaptive features. Int. J. Environ. Sci. Technol. 20, 12155–12166 (2023). https://doi.org/10.1007/s13762-022-04635-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-022-04635-5