Abstract
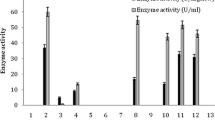
Amidase hydrolases/acyltransferases are of considerable industrial interest due to potential applications in the production of useful hydroxamic acids. The test strain, capable of acetamide degradation, was isolated by an enrichment technique with acetamide as sole source of nitrogen. Based on morphology, physiological tests and biochemical tests, this isolate was identified as Bacillus sp. and on the basis of 16S Ribosomal ribonucleic acid sequence, a phylogenetic tree was drawn and was identified as Bacillus megaterium. Resting cells containing active acyltransferase enzyme were prepared and immobilized in the gel beads of sodium alginate, agar, polyacrylamide and polyvinyl alcohol–alginate. The beads were tested for acyltransferase using Iron (III) chloride reagents at 55°C and were found to be affected by substrate concentration, type of buffer, buffer pH and reaction temperature. These factors were optimized using sodium alginate immobilized beads. This study proved useful in understanding the technique of immobilization of acyltransferase enzyme, its operational stability and its importance in the synthesis of hydroxamic acid.









Similar content being viewed by others
References
Asano Y, Tashibana M, Tani Y, Yamada H (1982) Aliphatic nitrile hydratase from Arthrobacter sp. J1: purification and characterization. Agric Biol Chem 46(5):1165–1174
Banerjee A, Sharma R, Banerjee V (2002) The nitrile degrading enzymes: current status and future prospects. Appl Microbol Biotechnol 60(1–2):33–44
Bhalla T, Kumar J, Kumar H, Agarwal H (1997) Amidase production by Rhodococcus sp. NHB-2. Natl Acad Sci Lett 20:139–142
Brammar W, Clarke H (1964) Induction and repression of Pseudomonas aeroginosa amidase. J Gen Microbiol 37:307–319
Brandao PFB, Clapp JP, Bull AT (2003) Diversity of nitrile hydratase and amidase enzyme genes in Rhodococcus erythropolis recovered from geographically distinct habitats. Appl Environ Microbiol 69(10):5754–5766
Chand D, Bhalla T, Kumar D, Vitzthum F (2004) Treatment of simulated wastewater containing toxic amides by immobilized Rhodococcus rhodochrous NHB-2 using a highly compact 5-stage plug flow rector. World J Microbiol Biotechnol 20(7):679–686
Cramp R, Gilmour M, Cowan DA (1997) Novel thermophilic bacteria producing nitrile-degrading enzymes. Microbiology 143:2313–2320
Fournand D, Pirat JL, Bigey F, Arnaud A, Galzy P (1997) Spectrophotometric assay of aliphatic monohydroxamic acids and α-, β- and γ-aminohydroxamic acids in aqueous medium. Anal Chem Acta 353(2–3):359–366
Fournand D, Bigey F, Arnaud A (1998) Acyl transfer activity of an amidase from Rhodococcus sp. strain R312: formation of a wide range of hydroxamic acids. Appl Environ Microbiol 64(8):2844–2852
Giorno L, Drioli E (2000) Biocatalytic membrane reactors: applications and perspectives. Trends Biotechnol 18(8):339–349
Hirrlinger B, Stolz A, Knackmuss H (1996) Purification and characterization of an amidase from Rhodococcus erythropolis MP50 which enantioselectively hydrolyzes 2-aryl propionamides. J Bacteriol 178(12):3501–3507
Jeanne M (1998) Properties of amides. Encyclopedia of occupational Health and Safety. Int Labour Office 4(104):73–79
Kao CM, Chen KF, Liu JK, Chou SM, Chen SC (2006) Enzymatic degradation of nitriles by Klebsiella oxytoca. Appl Microbiol Biotechnol 71(2):228–233
Kashiwagi M, Fuhshuku K, Sugai T (2004) Control of the nitrile-hydrolyzing enzyme activity in Rhodococcus rhodochrous IFO 15564: preferential action of nitrile hydratase and amidase depending on the reaction condition factors and its application to the one_pot preparation of amides from aldehydes. J Mol Catal B: Enzym 29(1–6):249–258
Kierstan M, Bucke C (1977) The immobilization of microbial cells, subcellular organelles and enzymes in calcium alginate gels. Biotechnol Bioeng 19(3):387–397
Kierstan MPJ, Coughlan MP (1985) Immobilization of cells and enzymes by gel entrapment. In: Woodward J (ed) Immobilized cells and enzymes. IRL Press, Oxford, pp 43–45
Kohyama E, Dohi M, Yoshimura A, Yoshida T, Nagasawa Toru (2007) Remaining acetamide in acetonitrile degradation using nitrile hydratase- and amidase-producing microorganisms. Appl Microbiol Biotechnol 74(4):829–835
Kotlova E, Chestukhina G, Astaurova O, Leonova T, Yanenko A, Debabov V (1999) Isolation and primary characterization of an amidase from Rhodococcus rhodochrous M8. Biochemistry 64(4):384–389
Kubac D, Kaplan O, Elisakova V, Patek M, Vejvoda V, Slamova K, Tothova A, Lemaire M, Gallienne E, Lutz-Wahl S, Fischer L, Kuzma M, Pelantova H, van Pelt S, Bolte J, Kren V, Martinkova L (2008) Biotransformation of nitriles to amides using soluble and immobilized nitrile hydratase from Rhodococcus erythropolis A4. J Mol Catal B: Enzym 50(2–4):107–113
Kumar H, Prasad S, Raj J, Bhalla TC (2006) Constitutive acetonitrile hydrolyzing activity of Nocardia globerula NHB-2: optimization of production and reaction conditions. Ind J Exp Biol 40:240–245
Landis WG, Ming-Ho Y (2003) Introduction to environmental toxicology—impacts of chemicals upon ecological systems, 3rd edn. Lewis Publishers, New York, p 242
Layh N, Stolz A, Forster S, Effenberger F, Knackmuss HJ (1992) Enantioselective hydrolysis of O-acetylmandelonitrile to O-acetylmandelic acid by bacterial nitrilases. Arch Microbiol 158(6):405–411
Martinkova L, Kren V, Vejvoda V (2008) Selection and screening for enzymes of nitrile metabolism. J Biotechnol 133(3):318–326
Mata JA, Martinez-Canovas J, Quesada E, Bejar V (2002) A detailed phenotypic characterisation of the type strains of Halomonas sp. Syst Appl Microbiol 25(3):360–375
Mateo C, Palomo JM, van Langen LM, van Rantwijk F, Sheldon RA (2004) A new, mild cross-linking methodology to prepare cross-linked enzyme aggregates. Biotehnol Bioeng 86(3):273–276
Mayaux J, Carbeland E, Solution F, Faucher D, Petre D (1990) Purification, cloning and primary structure of an enantiomer selective amidase from Brevibacterium sp. R312: structural evidence for genetic coupling with nitrile hydratase. J Bacteriol 172(12):6764–6773
Nawaz MS, Khan AA, Seng JE, Leakey JE, Siltonen PH, Cerniglia CE (1994) Purification & characterization of an amidase from an acryl amide degrading Rhodococcus sp. J Appl Microbiol 60(9):3343–3348
Nawaz MS, Billedeau SM, Cerniglia CE (1998) Influence of selected physical parameters on the biodegradation of acrylamide by immobilized cells of Rhodococcus sp. Biodegradation 9(5):381–387
Pal A, Samanta TB (1999) β-Lactamase-free penicillin amidase from Alcaligenes sp. Isolation strategy, strain characteristics, and enzyme immobilization. Curr Microbiol 39(5):244–248
Pattanapipitpaisal P, Brown NL, Macaskie LE (2001) Chromate reduction by Microbacterium liquefaciens immobilized in polyvinyl alcohol. Biotechnol Lett 23(1):61–65
Schoevaart R, Wolbers MW, Golubovic M, Ottens M, Kieboom AP, van Rantwijk F, van der Wielen LA, Sheldon RA (2004) Preparation, optimization, and structures of cross-linked enzyme aggregates (CLEAs). Biotechnol Bioeng 87(6):754–762
Ventosa A, Quesada E, Rodriguez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A (1982) Numerical taxonomy of moderately halophilic Gram-negative rods. J Gen Microbiol 128(9):1959–1968
Wiegand S, Steffen M, Koch R (1999) Isolation and identification of microorganisms able to grow on the polyester amide BAK 1095. J Environ Polym Degrad 7(3):145–156
Yousefi D, Khodadadi A, Ganjidoust H, Badkoubi A (2009) Isolation and characterization of a novel native Bacillus strain capable of degrading diesel fuel. J Environ Sci Tech 6(3):435–442
Acknowledgments
This work was supported by Jaipur Engineering College and Research Centre (JECRC, Jaipur, India). Authors are thankful to Mr. Arpit Agarwal (Director, JECRC), Prof. Reena Mathur (HOD, Dept. of Zoology, University of Rajasthan, Jaipur), Dr. C.P. Malik from Jaipur National University, India and Dr. S.P. Chaurasia, Chem. Eng Dept., MNIT, Jaipur for their kind suggestions and helpful discussion. Authors also wish to thank Prof. Santra of Therachem Medilab, Jaipur (India) for sample purification by preparative HPLC and NCCS, Pune (India) for gene sequencing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sogani, M., Mathur, N., Bhatnagar, P. et al. Biotransformation of amide using Bacillus sp.: isolation strategy, strain characteristics and enzyme immobilization. Int. J. Environ. Sci. Technol. 9, 119–127 (2012). https://doi.org/10.1007/s13762-011-0005-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-011-0005-7